Download Curated Microbial Reference Database
The Download Curated Microbial Reference Database tool downloads selected references as sequence lists and/or indexes that can be used with downstream analysis tools.
To run the tool, go to:
Tools | Microbial Genomics Module (![]() ) | Metagenomics (
) | Metagenomics (![]() ) | Databases (
) | Databases (![]() ) | Taxonomic Analyses (
) | Taxonomic Analyses (![]() ) | Download Curated Microbial Reference Database (
) | Download Curated Microbial Reference Database (![]() )
)
In the first window (figure 16.1), select the database you wish to download.
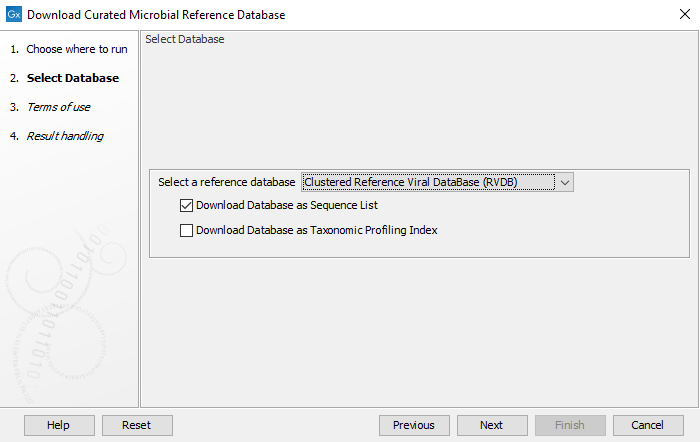
Figure 16.1: Select the database and output format
You can choose between several databases:
The sizes of the different formats are indicated below each entry as "size (of) sequence list/taxpro index/metagenome index", respectively. A star indicates that the format is not available for that item.
- QMM-H. QIAGEN Microbial Metagenome - Human Host database is a comprehensive microbial reference database for classification of whole metagenome data with Classify Whole Metagenome Data. The database is based on all chromosome level or complete RefSeq genomes of archaea, bacteria, viruses, protozoa and fungi, and UniVec_Core sequences. Genome sequences and annotations are from Genbank (https://www.ncbi.nlm.nih.gov/genbank/). UniVec_Core sequences are from the UniVec Database (http://www.ncbi.nlm.nih.gov/tools/vecscreen/univec/). Human genome assemblies hg38 (GCA_000001405.29) and T2T (GCA_009914755.4) are included as the host reference.
Size: 90.3 GB/*/89.5 GB. - QMI-PTDB Genus. QIAGEN Microbial Insights - Prokaryotic Taxonomy Database is a microbial reference database for taxonomic profiling of bacteria and archaea. The database represents all genera with a varying number of species per genus.
Genome sequences and annotations are from the NCBI Reference Sequence Database (RefSeq; https://www.ncbi.nlm.nih.gov/refseq/) and have been annotated with taxonomy from the Genome Taxonomy Database (GTDB; https://gtdb.ecogenomic.org).
The database was created by selecting one representative genome per species, and subsequently reducing the relative number of species per genus to meet the desired database size. For reduction, higher assembly status, lower number of contigs, and longer total length was prioritized. All genomes marked as "reference genome" were retained. So were species commonly included in microbial reference standards.
When running Taxonomic Profiling with the QMI-PTDB Genus database, 32GB of memory is required.
Size: 15.7 GB/22.4 GB/*. - QMI-PTDB Family. QIAGEN Microbial Insights - Prokaryotic Taxonomy Database is a microbial reference database for taxonomic profiling of bacteria and archaea. The database represents all families with a varying number of genera per family.
Genome sequences and annotations are from the NCBI Reference Sequence Database (RefSeq; https://www.ncbi.nlm.nih.gov/refseq/) and have been annotated with taxonomy from the Genome Taxonomy Database (GTDB; https://gtdb.ecogenomic.org).
The database was created by selecting one representative genome per genus, and subsequently reducing the relative number of genera per family to meet the desired database size. For reduction, higher assembly status, lower number of contigs, and longer total length was prioritized. All genomes marked as "reference genome" were retained. So were species commonly included in microbial reference standards.
When running Taxonomic Profiling with the QMI-PTDB Family database, 16GB of memory is recommended.
Size: 4.6 GB/6.5 GB/*. - MGnify Unified Human Gastrointestinal Genome (UHGG). A database of metagenomic-assembled genomes from human gut samples, curated and hosted by MGnify [Gurbich et al., 2023], EMBL-EBI (https://www.ebi.ac.uk/metagenomics/browse/genomes).
Size: 3.5 GB/7.8 GB/*. - MGnify Chicken Gut. A database of metagenomic-assembled genomes from chicken gut samples, curated and hosted by MGnify [Gurbich et al., 2023], EMBL-EBI (https://www.ebi.ac.uk/metagenomics/browse/genomes).
Size: 1.0 GB/2.1 GB/*. - MGnify Pig Gut. A database of metagenomic-assembled genomes from pig gut samples, curated and hosted by MGnify [Gurbich et al., 2023], EMBL-EBI (https://www.ebi.ac.uk/metagenomics/browse/genomes).
Size: 1.0 GB/2.0 GB/*. - Unclustered Reference Viral DataBase (U-RVDB). Unclustered Reference Viral Database for virus detection [Goodacre et al., 2018]. The database includes curated viral, virus-related and virus-like nucleotide sequences except bacterial viruses, which are excluded.
Size: 42.0 GB/*/1.6 GB. - Clustered Reference Viral DataBase (C-RVDB). Clustered Reference Viral Database for virus detection [Goodacre et al., 2018]. The database includes curated viral, virus-related and virus-like nucleotide sequences except bacterial viruses, clustered at 98% sequence similarity.
Size: 0.6 GB/2.7 GB/1.27 GB.
When you have made your database selection, choose which format you wish to download.
- Download Database as Sequence list. Produces an annotated sequence list.
- Download Database as Whole Metagenome Index. This index type is used by the Classify Whole Metagenome Data tool (Classify Whole Metagenome Data).
- Download Database as Taxonomic Profiling Index. This index type is used by e.g., the Taxonomic Profiling tool (Taxonomic Profiling).
Some of the databases offered are derived work, licensed under a Creative Commons Attribution-ShareAlike (CC BY-SA) license. We offer free access to those without requiring a CLC product license. They can be downloaded using the CLC Genomics Workbench with the Microbial Genomics Module installed in Viewing Mode. The downloaded files can then be exported to non-proprietary formats.
