De Novo Assemble Metagenome parameters
To run the tool, go to:
Tools | Microbial Genomics Module (![]() ) | Metagenomics (
) | Metagenomics (![]() ) | De Novo Assemble Metagenome (
) | De Novo Assemble Metagenome (![]() )
)
Select the sequence lists or single sequences to assemble.
Set assembly parameters (figure 4.1).
- Minimum contig length. Contigs below this length will not be reported. For very complex datasets containing reads from many closely related species, the assembler will often produce shorter contigs. For such cases, it is recommended to set a lower threshold in order to cover a larger proportion of the metagenome with contigs. Reversely, for metagenomes of low complexity, it is often wise to set a higher threshold in order to avoid duplication.
- Execution mode
- Fast. The assembler is iterated once with a predifined wordsize (
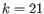 ).
).
- Longer contigs. the assembler is iterated three times with increasing wordsize (
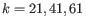 ), using the contigs from the previous iteration as input in the next iteration together with the input reads.
), using the contigs from the previous iteration as input in the next iteration together with the input reads.
- Fast. The assembler is iterated once with a predifined wordsize (
- Perform scaffolding. If selected, as the last step of the assembly process the assembler attempts to join contigs using paired-end information. Since paired-end information is needed to perform scaffolding, this option is disabled for single-end sequences.
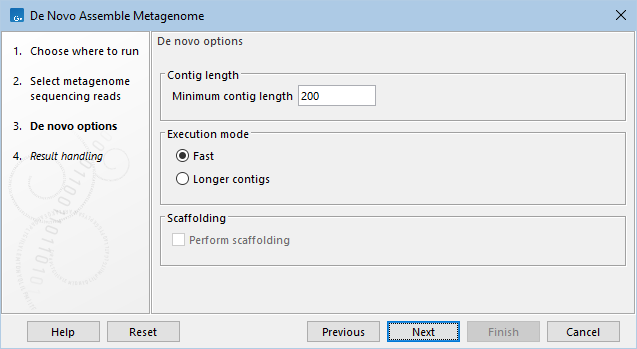
Figure 4.2: Setting parameters for the assembly.
