Annotate CDS with Best DIAMOND Hit
Annotate CDS with Best DIAMOND Hit allows you to annotate a set of contigs containing CDS annotations with their best DIAMOND hit. This tool is particularly useful for large data sets, as an alternative to Annotate CDS with Best BLAST Hit.
DIAMOND is a sequence aligner for protein and translated DNA searches, designed for high performance analysis of big sequence data, see https://github.com/bbuchfink/diamond. The key features are:
- Pairwise alignment of proteins and translated DNA at 500x-20,000x speed of BLAST.
- Frameshift alignments for long read analysis.
- Low resource requirements and suitable for running on standard desktops or laptops.
To start the analysis, go to:
Tools | Microbial Genomics Module (![]() ) | Functional Analysis (
) | Functional Analysis (![]() ) | Annotate CDS with Best DIAMOND Hit (
) | Annotate CDS with Best DIAMOND Hit (![]() )
)
Select the CDS-annotated contigs to be annotated with DIAMOND hits.
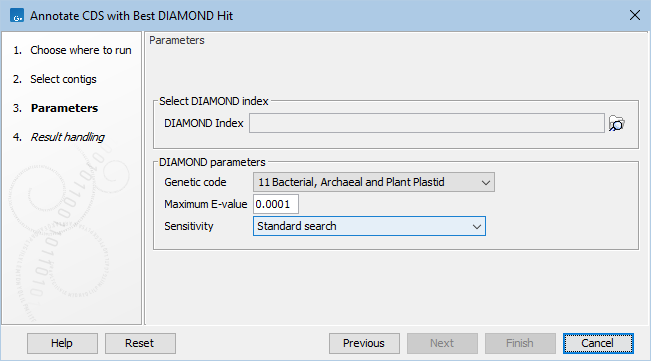
Figure 12.9: Annotate CDS with Best DIAMOND Hit parameters.
In the Parameters dialog page (figure 12.9), set the following
- DIAMOND Index. Select the relevant indexes.
Indexes can be generated by downloading a database with the Download Protein Database tool (Download Protein Database) and building and index using the Create DIAMOND Index tool (Create DIAMOND Index).
- Genetic code. The genetic code used for translating CDS to proteins.
- Maximum E-value. Maximum expectation value (E-value) threshold for saving hits.
- Sensitivity: Select DIAMOND sensitivity:
- Faster search: The fastest search
- Fast search: Designed for finding hits of >90% identity
- Standard search: Designed for finding hits of >60% identity
- Mid-sensitive search: More sensitive than standard search and faster than sensitive search.
- Sensitive search: Designed for finding hits of >40% identity
- More sensitive search: Designed for finding hits of >40% identity with some motif masking disabled
- Very sensitive search: Designed for finding hits of 40% identity
- Most sensitive search: The most sensitive search
The tool will output a copy of the input file with the DIAMOND Hit annotations. The tool can also output an annotation table summarizing information about the annotations added to the sequence list. Finally it is possible to generate a report containing information about the input file, the DIAMOND database and the amount of CDS annotated with a DIAMOND hit.
If a DIAMOND index was created from a protein sequence list containing metadata (such as GO terms or taxonomy information), the original metadata will be transferred to the annotations created by this tool.
