Analyze Viral Hybrid Capture Panel Data
This template workflow has been replaced by Analyze QIAseq xHYB Viral Panel Data (Human host), see Analyze QIAseq xHYB Viral Panel Data (Human host).
The Analyze Viral Hybrid Capture Panel Data (legacy) template workflow is designed for detecting viruses, calculating abundances, and for calling variants for viruses identified from data generated using hybrid capture panels. The workflow performs read trimming, creates a QC report, cleans the dataset of host DNA, calculates viral abundances, and maps the reads to the most abundant viral reference for variant calling.
If your panel does not contain control genes, the workflow should be modified by right clicking on the workflow in the tool box and opening a copy of the workflow. Remove the Map Reads to Human Control Genes workflow element plus its input and output, and save the modified workflow. When you run the modified workflow, human control genes are no longer required.
Preliminary steps to run the Analyze Viral Hybrid Capture Panel Data (legacy) workflow
Before starting the workflow,
- Download reference viral genomes using either the Download Custom Microbial Reference Database (see the Working with databases chapter), the viral databases from the Download Curated Microbial Reference Database tool or create a database using the Update Sequence Attributes in Lists tool (https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Update_Sequence_Attributes_in_Lists.html)
- Create a taxonomic profiling index for calculating abundance (see Create Taxonomic Profiling Index).
To run the workflow, go to:
Toolbox | Legacy Tools (![]() ) |Analyze Viral Hybrid Capture Panel Data (legacy) (
) |Analyze Viral Hybrid Capture Panel Data (legacy) (![]() )
)
- Specify the sample(s) or folder(s) of samples you would like analyze (figure 20.4) and click Next. Note that if you select several items, they will be run as batch units.
Figure 20.4: Select the reads from the sample(s) you would like to analyze - Specify the human control genes as a sequence list here (figure 20.5). Alternatively, if your panel does not contain control genes, the workflow should be modified by right clicking on the workflow in the tool box and opening a copy of the workflow. Then remove the Map Reads to Reference tool plus its input and output and save the modified workflow. When you run the modified workflow, human control genes are no longer required.
Figure 20.5: Select the human control genes or reference - Define batch units using organisation of input data to create one run per input or use a metadata table to define batch units. Click Next.
- The next wizard window gives you an overview of the samples present in the selected folder(s). Choose which of these samples you want to analyze in case you are not interested in analyzing all the samples from a particular folder (figure 20.6).
Figure 20.6: Choose which of the samples present in the selected folder(s) you want to analyze. - You can specify a trim adapter list and set up parameters if you would like to trim your sequences (figure 20.7).
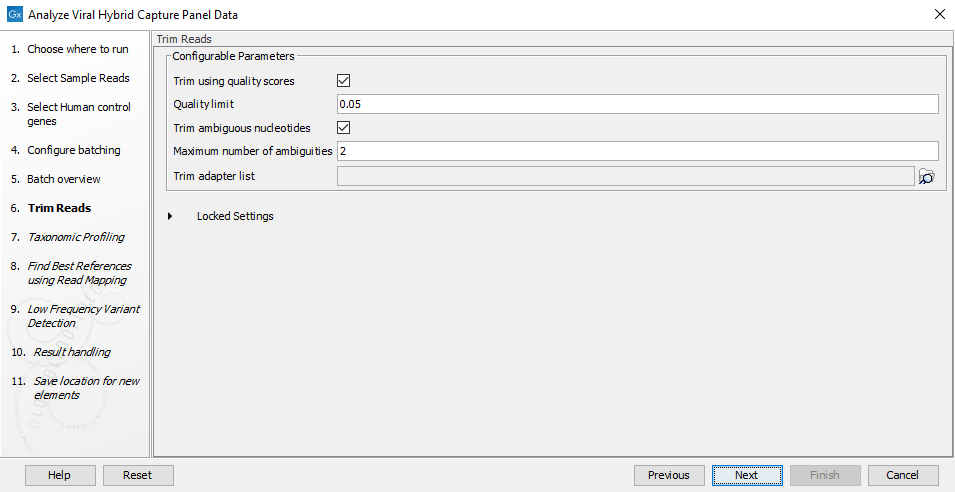
Figure 20.7: Choose trimming settings and optionally add an adapter trim list for trimming sequencing reads.The parameters that can be set are:
- Trim ambiguous nucleotides: if checked, this option trims the sequence ends based on the presence of ambiguous nucleotides (typically N).
- Maximum number of ambiguities: defines the maximal number of ambiguous nucleotides allowed in the sequence after trimming.
- Trim using quality scores: if checked, and if the sequence files contain quality scores from a base-caller algorithm, this information can be used for trimming sequence ends.
- Quality limit: defines the minimal value of the Phred score for which bases will not be trimmed.
- Trim adapter list: Specifying a trim adapter list is optional but recommended to ensure the highest quality data for your analysis (figure 20.7)
Learn about trim adapter lists at https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Trim_adapter_list.html)
- In the next wizard window "Taxonomic Profiling", select the viral reference database index you will use to map the reads (figure 20.8). It is also possible to "Filter host reads". You must then specify the index of the host genome (in the case of human virus, the Homo sapiens GRCh38 for example). Note that if your panel uses human control genes, a taxonomic profiling index of the human genome should be used as "Host index".
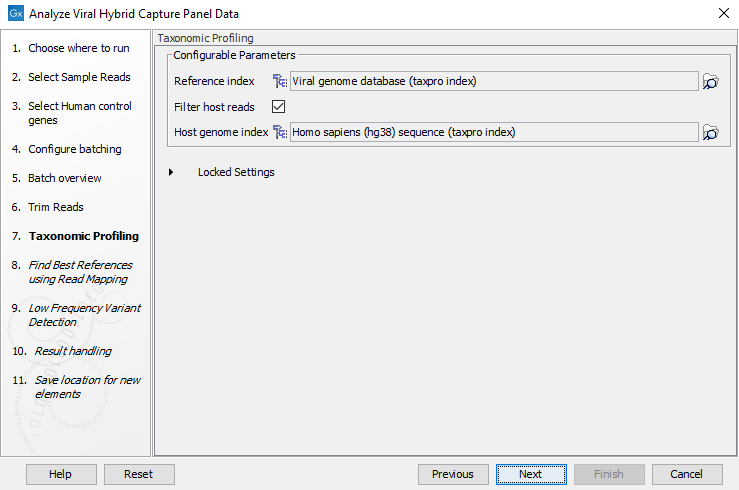
Figure 20.8: Select taxonomic profiling index - In the next wizard window, select the viral reference database you will use to find the best matching reference (figure 20.9). The best matching reference will be used for read mapping and variant calling. If you wish to have variant calls annotated with amino acid changes the input database should contain CDS annotations.
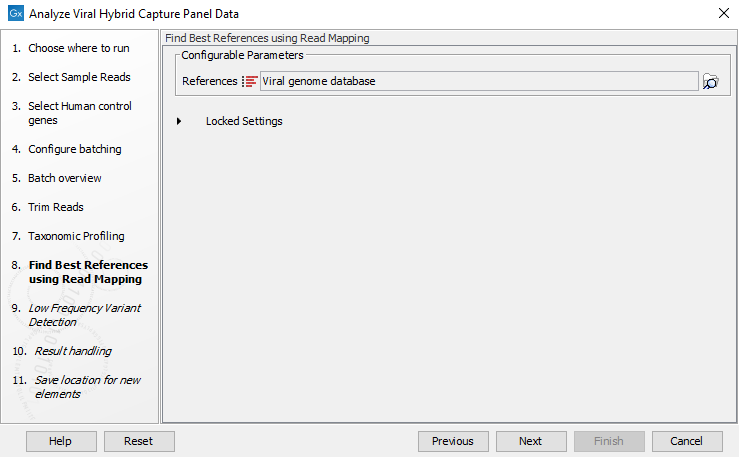
Figure 20.9: Select viral reference database - In the next wizard window, specify the parameters for the Low Frequency Variant Detection tool (figure 20.10). Note that variants are filtered after variant detection to coverage
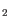 30x and frequency
30x and frequency 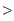 20%.
20%.
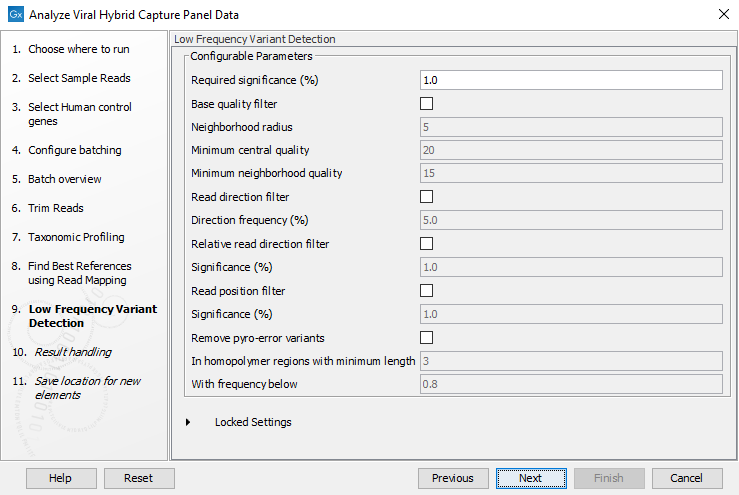
Figure 20.10: Specify the parameters to be used by the Low Frequency Variant Detection tool.The parameters that can be set are:
- Required significance: The required significance level for low frequency variant calls.
- Base quality filter: The base quality filter can be used to ignore the reads whose nucleotide at the potential variant position is of dubious quality.
- Neighborhood radius: Determine how far away from the current variant the quality assessment should extend.
- Minimum central quality: Reads whose central base has a quality below the specified value will be ignored. This parameter does not apply to deletions since there is no "central base" in these cases.
- Minimum neighborhood quality: Reads for which the minimum quality of the bases is below the specified value will be ignored.
- Read direction filter: The read direction filter removes variants that are almost exclusively present in either forward or reverse reads.
- Direction frequency %: Variants that are not supported by at least this frequency of reads from each direction are removed.
- Relative read direction filter: The relative read direction filter attempts to do the same thing as the Read direction filter, but does this in a statistical, rather than absolute, sense: it tests whether the distribution among forward and reverse reads of the variant carrying reads is different from that of the total set of reads covering the site. The statistical, rather than absolute, approach makes the filter less stringent.
- Significance %: Variants whose read direction distribution is significantly different from the expected with a test at this level, are removed. The lower you set the significance cut-off, the fewer variants will be filtered out.
- Read position filter: This filter removes variants that are located differently in the reads carrying it than would be expected given the general location of the reads covering the variant site.
- Remove pyro-error variants: This filter can be used to remove insertions and deletions in the reads that are likely to be due to pyro-like errors in homopolymer regions. There are two parameters that must be specified for this filter:
- In homopolymer regions with minimum length: Only insertion or deletion variants in homopolymer regions of at least this length will be removed.
- With frequency below: Only insertion or deletion variants whose frequency (ignoring all non-reference and non-homopolymer variant reads) is lower than this threshold will be removed.
- In the Result handling window, pressing the button Preview All Parameters allows you to preview - but not change - all parameters. Choose to save the results (we recommend to create a new folder for it) and click Finish.
The output will be saved in the folder you selected. An example of the output can be seen in figure 20.11.
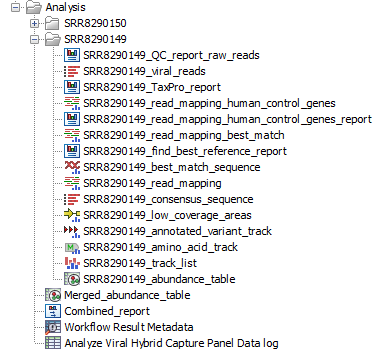
Figure 20.11: Output of analysis of viral hybrid capture panel data
The output generated for each sample is:
- QC report raw reads: QC report on the raw reads. Contains information on number of input reads, length, quality and nucleotide distributions.
- Viral reads: list of the sequences that were successfully trimmed and mapped to the best reference.
- TaxPro report: output from the Taxonomic Profiling tool. Contains information on number of reads mapping to the reference database and the host, if host filtering was enabled.
- Read mapping human control genes: mapping of the reads to the human control genes.
- Read mapping human control genes report: contains information on the read mapping to the selected controls such as number of reads mapped and read length distributions.
- Best reference report: contains information on "Best match" reference identified by Find Best References using Read Mapping, and the number of reads and unique reads mapped to this reference.
- Best match sequence: the "Best match" reference sequence as identified by the Find Best References using Read Mapping tool.
- Read mapping: reads mapped to the "Best match" viral reference. Output from Local Realignment.
- Consensus sequence: a consensus sequence generated from the read mapping. Consensus is not calculated in low coverage regions. These positions are instead replaced with Ns.
- Low coverage areas: track of regions of the best match reference genome with coverage < 30x.
- Trim report: report from the Trim Sequences tool (see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Trim_output.html).
- Abundance table: contains abundance for all detected viral species.
- Viral reads: read mapping to the viral reference database.
- Annotated variant track: output from the Low Frequency Variant Detection tool. Note that variants are post filtered to coverage
 30x and frequency
30x and frequency  20%.
20%.
- Amino acid track: only generated if the best match contains CDS annotations.
For each batch analysis run, the following outputs are generated:
- Combined report: combines the information from the output report including QC, taxonomic profiling and mapping reports.
- Merged abundance table: An table containing abundance for all input samples. See figure 20.12 for an example of an analysis run of two HPV samples.
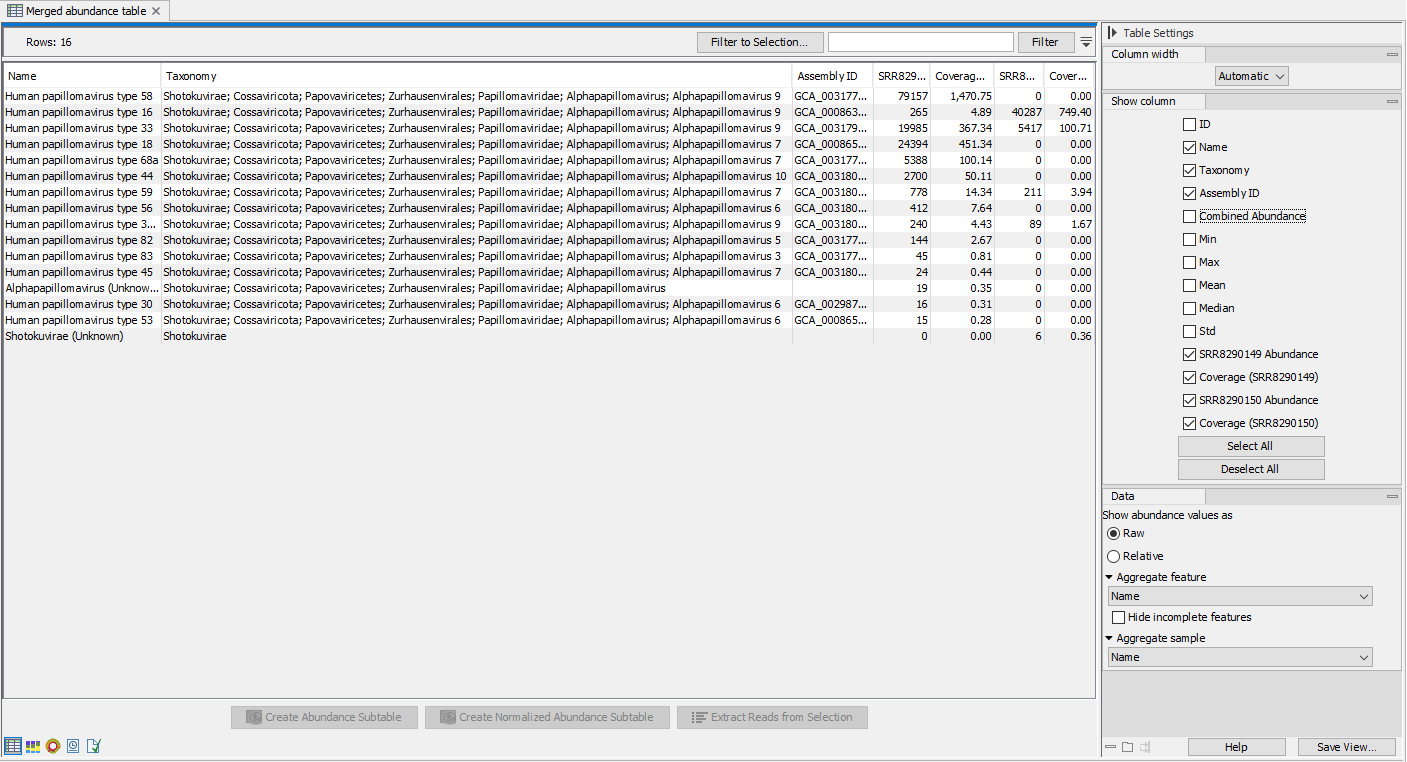
Figure 20.12: Merged abundance table from analysis of two HPV samplesMerged abundance tables can be used as input for various tools:
#18138#>
