Mapping SOLid reads in color space
The SOLiD sequencing technology from Applied Biosystems is different
from other sequencing technologies since it does not sequence one base
at a time. Instead, two bases are sequenced at a time in an
overlapping pattern. There are 16 different dinucleotides, but in the
SOLiD technology, the dinucleotides are grouped in four carefully
chosen sets, each containing four dinucleotides. The colors are as
follows:

Notice how a base and a color uniquely defines the following base. This approach can be used to deduce a whole sequence from the initial nucleotide and a series of colors. Here is a sequence and the corresponding colors.
![]()
The colors do not uniquely define the sequence. Here is another sequence with the same list of colors:
![]()
But if the first nucleotide is known, the colors do uniquely define the remaining sequence. This is exactly the strategy used in SOLiD sequencing: The first nucleotide is known from the primer used, and the remaining nucleotides are deduced from the colors.
As with other sequencing technologies, errors do occur with the SOLiD technology. If a single nucleotide is changed, two colors are affected since a single nucleotide is contained in two overlapping dinucleotides:
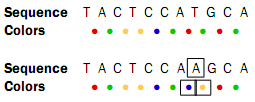
Sometimes, a wrong color is determined at a given position. Due to the dependence between dinucleotides and colors, this affects the remaining sequence from the point of the error:
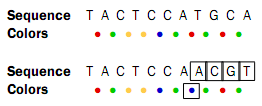
Thus, when the instrument makes an error while determining a color, the error mode is very different from when a single nucleotide is changed. This ability to differentiate different types of errors and differences is a very powerful aspect of SOLiD sequencing. With other technologies sequencing errors always appear as nucleotide differences.
Subsections
