The un-grouped sample
An example of an un-grouped annotated sample is shown in figure 28.31.
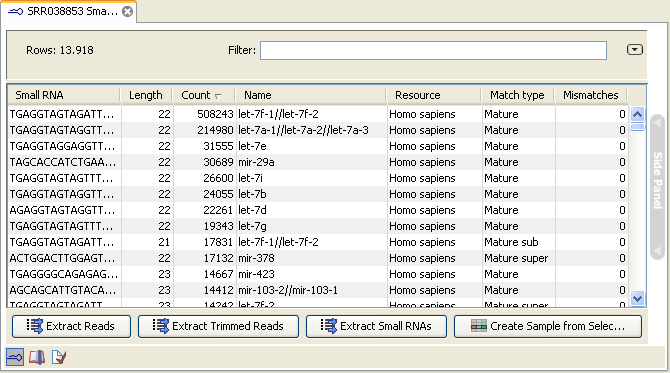
Figure 28.31: An ungrouped annotated sample.
By selecting one or more rows in the table, the buttons at the bottom of the view can be used to extract sequences from the table:
- Extract Reads (
 )
) - This will extract the original sequencing reads that contributed to this tag. Figure 28.32 shows an example of such a read. The reads include trim annotations (for use when inspecting and double-checking the results of trimming). Note that if these reads are used for read mapping, the trimmed part of the read will automatically be removed. If all rows in the sample are selected and extracted, the sequence list would be the same as the input except for the reads that did not meet the adapter trim settings and the sampling thresholds (tag length and number of copies).
- Extract Trimmed Reads (
 )
) - The same as above, except that the trimmed part has been removed.
- Extract Small RNAs (
 )
) - This will extract only one copy of each tag.
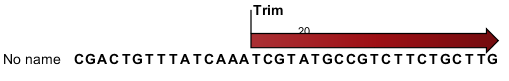
Figure 28.32: Extracting reads from a sample.
The button Create Sample from Selection (![]() ) can be used to create a new sample based on the tags that are selected. This can be useful in combination with filtering and sorting.
) can be used to create a new sample based on the tags that are selected. This can be useful in combination with filtering and sorting.
