Differences among the variants called by the three variant callers
The Variant Detection tools will call SNVs, MNVs (which are neighboring SNVs for which there is evidence in the data that they occur together), small to medium-sized insertions and deletions (the size of the insertions and deletions that the variant detection tools are able to call is restricted by the fact that they need to be represented within a single read), and replacements (which are neighboring SNVs and indels).As the tools differ in their underlying assumptions about the data, they differ in their assessments of when there is enough information in the data for a variant to be called, and hence will call different variants. However, when run with the same filter settings (Filters for a description of the filters), you will generally have that:
- The Basic Variant Caller will call the highest number of variants. It will also do this relatively quickly, as it does not do any error-model estimation.
- The Low Frequency Variant Caller will call a subset of the variants called by the Basic Variant caller. The variants called by the Basic Variant Caller that the Low Frequency Variant Caller will NOT call, are those that, according the error model that the Low Frequency Variant Caller estimates from the data, are likely to have been caused by sequencing errors. The Low Frequency Variant Caller will be the slowest of the three variant callers as it (1) estimates an error-model and (2) calls Low Frequency variants (and not just those that are in accordance with a specified ploidy model).
- The Fixed Ploidy Variant Caller will call a subset of the variants called by the Low Frequency Variant caller. The variants called by the Low Frequency Variant Caller that the Fixed Ploidy Variant Caller will NOT call, are those that, according to the assumed ploidy of the sample analyzed and the error model that the Fixed Ploidy Variant Caller estimates from the data, are likely to have been caused by either mapping errors or by sequencing errors.

Figure 21.46: The differences in variants called by the three variant callers. The variant callers have all been run with same the filter settings (those that are the defaults for the Low Frequency Variant Caller).
Figure 21.46 shows variant calls produced by the three variant callers when run with the same filter settings, more precisely those that are default for the Low Frequency Variant Caller. The numbers of called variants are shown in the left part of the figure, under the variant track names 'basicV2', 'LowFreq' and 'FixedV2'. The Basic Variant Caller calls most variants and the Fixed Ploidy the least. The Fixed Ploidy Variant Caller calls a subset of those called by the Low Frequency Variant caller, which in turn calls a subset of those called by the Basic Variant caller -- in spite of the fact that there are 9 variants in the Low Frequency variant track that are not in the Basic Variant track. Although those 9 variants are in fact not in the Basic Variant track, they are 'sub-variants' of variants in that track. The highlighted variants in the figure is an example of this: The Basic variant caller has called a heterozygous 2bp MNV. The Low Frequency variant caller has judged that one on the SNVs constituting this 2bp MNV is likely to be the result of sequencing errors, and has only called one of the SNVs.
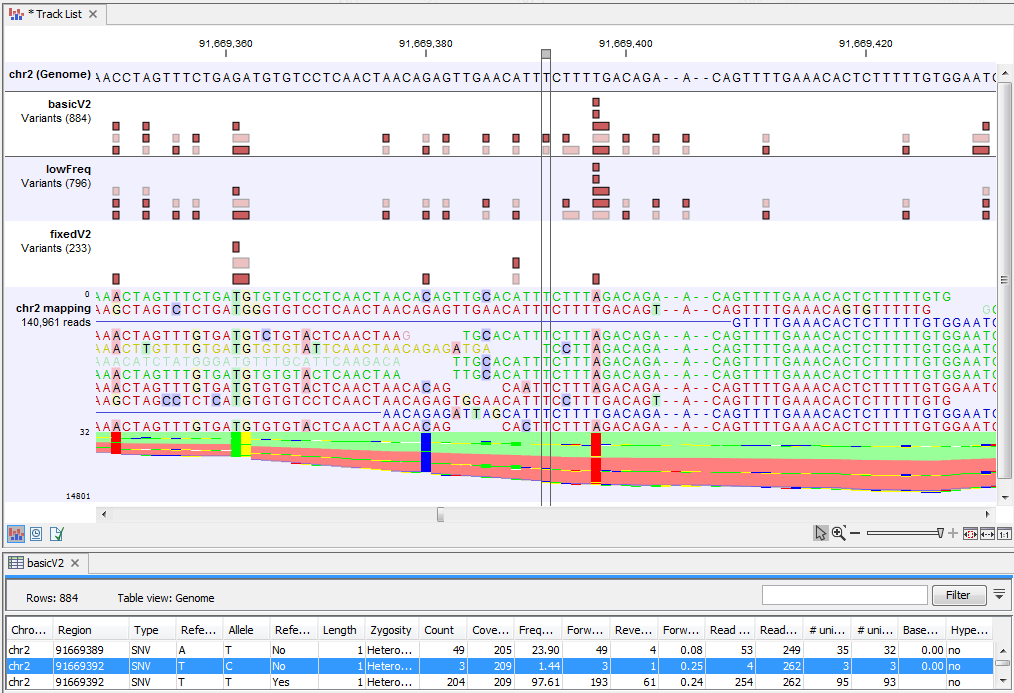
Figure 21.47: A variant is highlighted that is detected by the Basic Variant Caller but not by the Low Frequency or the Fixed Ploidy Variant Caller. The variant track for the Basic variant Caller variants is opened in the table-view at the bottom of the figure. The variant is present at a low frequency in a high coverage position, and is likely to have been caused by sequencing error.
In figure 21.47 a variant is highlighted that is detected by the Basic Variant Caller but not by the Low Frequency or the Fixed Ploidy Variant Caller. The variant is present at a low frequency in a high coverage position. The Low Frequency Variant Caller compares this evidence to the error model, and has decided that the three reads carrying the variant are likely to be the result of sequencing errors, rather than the result of a true variant. Figure 21.48 highlights a variant that is detected by both the Basic and the Low Frequency Variant Caller, but not the Fixed Ploidy. The variant is present at a higher frequency (14.22%) in a high coverage region (coverage 204). Observing the variant in 29 out of 204 reads is not likely to be due to sequencing errors. However, observing 29 reads from one allele and the remaining from the other in a diploid sample is highly unlikely, and the Fixed Ploidy Variant Caller judges that this variant is most likely caused by mapping errors (that is, a subset of the reads in the region being mapped there spuriously) and filters out this variant.
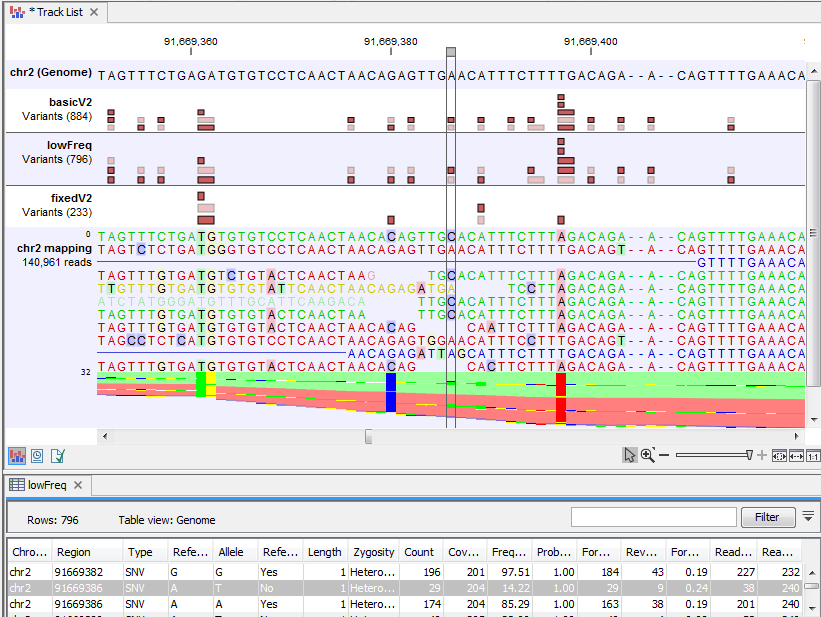
Figure 21.48: A variant is highlighted that is detected by the Basic and the Low Frequency but not by the Fixed Ploidy Variant Caller. The variant track for the Low Frequency Variant Caller variants is opened in the table-view at the bottom of the figure. The variant is present at a moderate frequency in a high coverage position, and is, under the assumed ploidy, most likely to have been caused by mapping error.
