Import Primer Pairs
The Import Primer Pairs importer can import descriptions of primer locations from a generic text format file or from a QIAGEN gene panel primer file. The primer location file describes the location of primers used for targeted resequencing and is used for primer trimming by the tool Trim Primers of Mapped Reads. This tool is particularly useful for trimming off primers when you have targeted data with overlapping reads.
The Import Primer Pairs can be found in the toolbar:
Import (![]() ) | Import Primer Pairs (
) | Import Primer Pairs (![]() )
)
This will open the wizard shown in figure 6.16. The first step is to select the data to import.
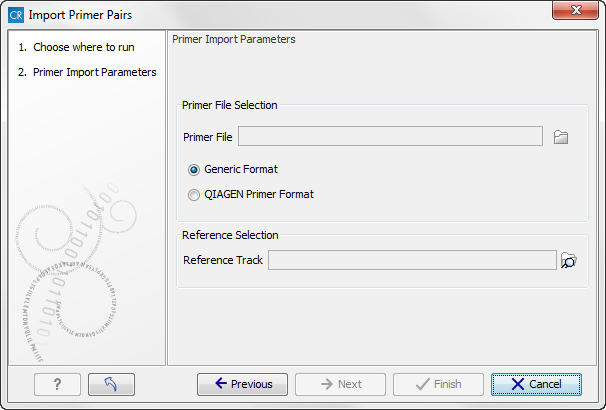
Figure 6.16: Select files to import.
- Primer File Click on the folder icon in the right side to select your primer pair location file.
There are two primer pair formats that can be imported by the Workbench.
- Generic Format Select this option for primer location files with the exception of QIAGEN gene panel primers. Provide your primer location information in a tab delimited text file with the following columns
- Column 1: reference name
- Column 2: primer1 first position on reference
- Column 3: primer1 last position on reference
- Column 4: primer2 first position on reference
- Column 5: primer2 last position on reference
- Column 6: amplicon name
An example of the format expected for each row is:
chr1 42 65 142 106 Amplicon1 - QIAGEN Primer Format Use this option for importing information about QIAGEN gene panel primers.
- Generic Format Select this option for primer location files with the exception of QIAGEN gene panel primers. Provide your primer location information in a tab delimited text file with the following columns
- Reference Track Use folder icon in the right side to select the relevant reference track.
Click on the button labeled Next to go to the wizard step choose to save the imported primer location file.
