Export graph data points to a file
Data points for graphs displayed along the sequence or along an alignment, mapping or BLAST result, can be exported to a semicolon-separated text file (csv format). An example of such a graph is shown in figure 6.36. This graph shows the coverage of reads of a read mapping (produced with CLC Genomics Workbench).

Figure 6.36: A graph displayed along the mapped reads. Right-click the graph to export the data points to a file.
To export the data points for the graph, right-click the graph and choose Export Graph to Comma-separated File. Depending on what kind of graph you have selected, different options will be shown: If the graph is covering a set of aligned sequences with a main sequence, such as read mappings and BLAST results, the dialog shown in figure 6.37 will be displayed. These kinds of graphs are located under Alignment info in the Side Panel. In all other cases, a normal file dialog will be shown letting you specify name and location for the file.
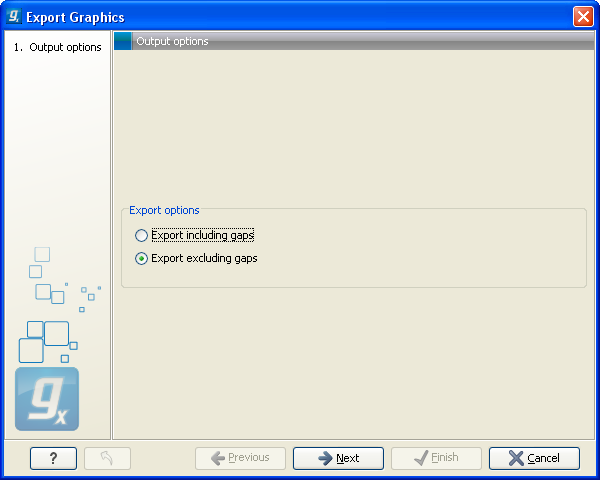
Figure 6.37: Choosing to include data points with gaps
In this dialog, select whether you wish to include positions where the main sequence (the reference sequence for read mappings and the query sequence for BLAST results) has gaps. If you are exporting e.g. coverage information from a read mapping, you would probably want to exclude gaps, if you want the positions in the exported file to match the reference (i.e. chromosome) coordinates. If you export including gaps, the data points in the file no longer corresponds to the reference coordinates, because each gap will shift the coordinates.
Clicking Next will present a file dialog letting you specify name and location for the file.
The output format of the file is like this:
"Position";"Value"; "1";"13"; "2";"16"; "3";"23"; "4";"17"; ...
