Viewing color space information
Importing data from SOLiD systems (see Importing SOLiD data) will from CLC Genomics Workbench version 3.1 be imported as color space. This means that if you open the imported data, it will look like figure 25.20
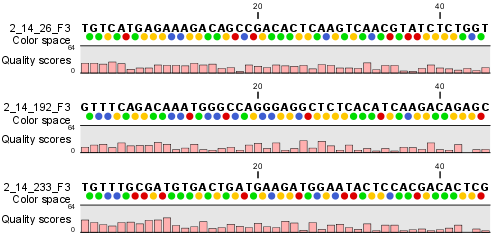
Figure 25.20: Color space sequence list.
In the Side Panel under Nucleotide info, you find the Color space encoding group which lets you define a few settings for how the colors should appear. These settings are also found in the side panel of mapping results and single sequences.
- Infer encoding
- This is used if you want to display the colors for non-color space sequence (e.g. a reference sequence). The colors are then simply inferred from the sequence.
- Show corrections
- This is only relevant for mapping results - it will show where the mapping process has detected color errors. An example of a color error is shown in figure 25.21.
- Hide unaligned ends
- This option determines whether color for the unaligned ends of reads should be displayed. It also controls whether colors should be shown for gaps. The idea behind this is that these color dots will interfere with the color alignment, so it is possible to turn them off.
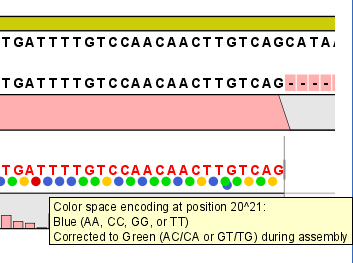
Figure 25.21: One of the dots have both a blue and a green color. This is because this color has been corrected during mapping. Putting the mouse on the dot displays the small explanatory message.
