Map reads to contigs
The "Map reads to contigs" tool allows mapping of reads to contigs. This can be relevant in situations such as when:
- Contigs have been imported from an external source
- The output from a de novo assembly is contigs with no read mapping
- You wish to map a new set of reads or a subset of reads to the contigs
Hence, in any situation where the reference of a mapping is contigs, the "Map reads to contigs" tool can be useful. The "Map reads to contigs" tool is similar to the "Map reads to Reference" tool in that both tools make use of the same read mapper and accept the same input reads. The main difference between the two tools is the output. The output from the "Map reads to contigs" tool is a de novo object that can be edited, which is in contrast to the reference sequence used when mapping reads to a reference.
To run the "Map reads to contigs" tool:
Toolbox | De Novo Sequencing (![]() ) | Map Reads to Contigs (
) | Map Reads to Contigs (![]() )
)
This opens up the dialog in figure 29.23.
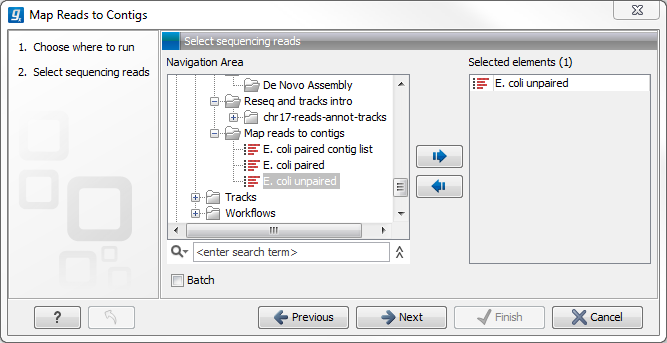
Figure 29.23: Select reads. The contigs will be selected in the next step.
The next step is to select the contigs to map the reads against (figure 29.24). Under "Contig masking", specify whether to include or exclude specific regions (for a description of this see Including or excluding regions (masking)). The contigs can be updated as part of the "Map Reads to Contigs" tool by selecting "Update contigs" at the bottom of the wizard. The advantage of using the read mapping in "Map Reads to Contigs" tool to update the contigs is that the read mapper is better than the de novo assembler at handling errors in reads.
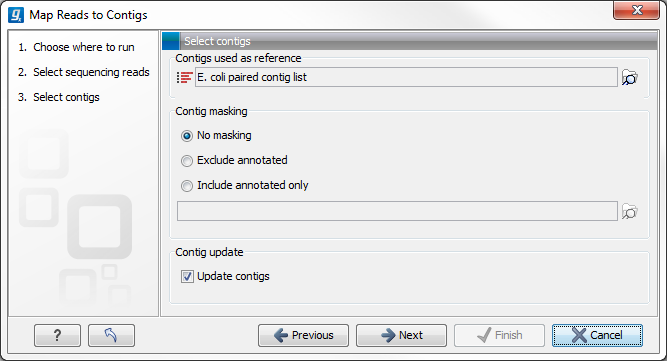
Figure 29.24: Select contigs and specify whether to use masking and the "Update contigs" function.
The next wizard steps are identical to the steps found in the "Map Reads to Reference" tool. For a description of these steps, please see Mapping parameters).
The output from the "Map Reads to Contigs" tool depends on whether tracks or stand-alone read mappings were selected in the last dialog. When stand-alone read mappings have been selected as output, it is possible to edit and delete in the contig sequences. Figure 29.25 shows the result of using "Map Reads to Reference" (top) and "Map Reads to Contigs" (bottom) on the exact same reads and contigs as input. Contig 1 from both analyses have been opened from their respective Contig Tables. The differences are highlighted with red arrows. Note that the output from the "Map Reads to Contigs" do not have a consensus sequence as the Contig itself will be the consensus sequence if "Update contigs" was selected.
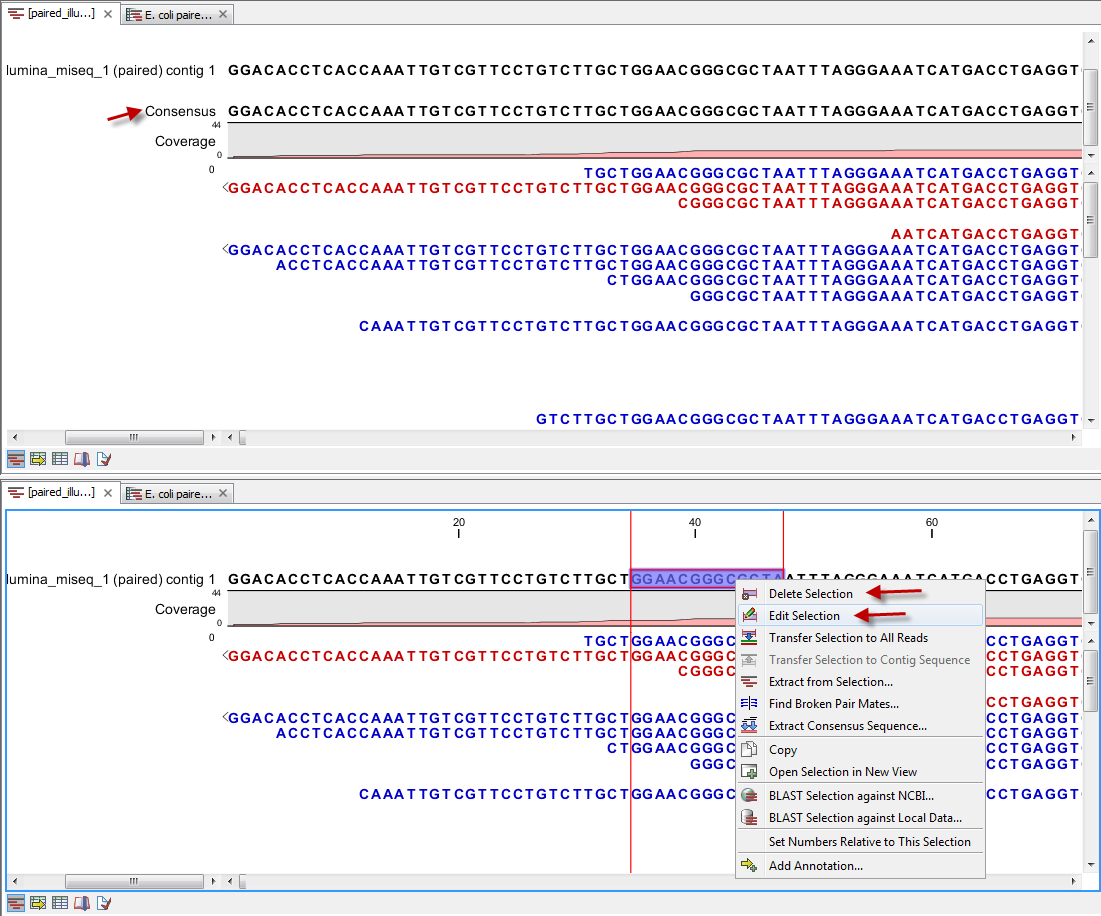
Figure 29.25: Two different read mappings performed with "Map Reads to Reference" (top) and "Map Reads to Contigs" (bottom). The differences are highlighted with red arrows.
By selecting "Update contigs" at the bottom of the wizard, the contigs generated by the de novo assembly are used as references that the reads used for the assembly input are mapped back to. The contigs themselves are updated based on the mapping results of the "Map Reads to Contigs". One advantage of using the read mapping in "Map Reads to Contigs" tool to update the contigs is that the read mapper is better than the de novo assembler at handling errors in reads. Specifically, the actions taken when contigs are updated are:
- Regions of a contig reference, where no reads map, are removed. This leads to the surrounding regions of the contig to be put together as one (figure 29.25).
- In the case of locations where reads map to a contig reference, but there are some mismatches to that contig, the contig sequence is updated to reflect the majority base at that location among the reads mapped there. If more than half of the reads contain a gap at that location, the contig sequence will be updated to include the gap.

Figure 29.26: When selecting "Update Contig" in the wizard, contigs will be updated according to the reads. This means that regions of a contig where no reads map will be removed.
