Use of multiple alignments
Once a multiple alignment is constructed it can form the basis for a number of analyses:- The phylogenetic relationship of the sequences can be investigated by tree-building methods based on the alignment.
- Annotation of functional domains, which may only be known for a subset of the sequences, can be transferred to aligned positions in other un-annotated sequences.
- Conserved regions in the alignment can be found which are prime candidates for holding functionally important sites.
- Comparative bioinformatical analysis can be performed to identify functionally important regions.
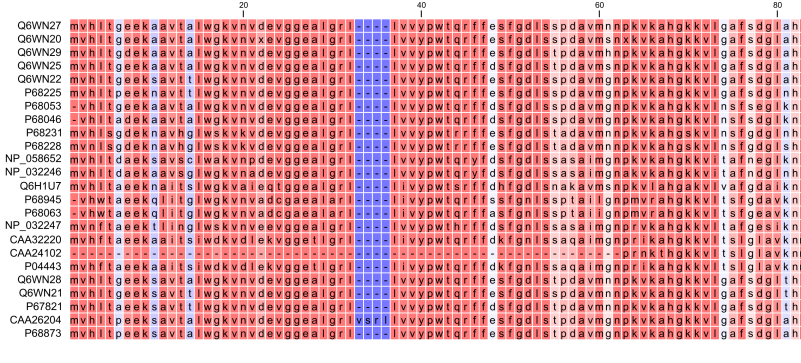
Figure 20.16: The tabular format of a multiple alignment of 24 Hemoglobin protein sequences. Sequence names appear at the beginning of each row and the residue position is indicated by the numbers at the top of the alignment columns. The level of sequence conservation is shown on a color scale with blue residues being the least conserved and red residues being the most conserved.
