Use of scoring matrices
Deciding which scoring matrix you should use in order of obtain the best alignment results is a difficult task. If you have no prior knowledge on the sequence the BLOSUM62 is probably the best choice. This matrix has become the de facto standard for scoring matrices and is also used as the default matrix in BLAST searches. The selection of a "wrong" scoring matrix will most probable strongly influence on the outcome of the analysis. In general a few rules apply to the selection of scoring matrices.
- For closely related sequences choose BLOSUM matrices created for highly similar alignments, like BLOSUM80. You can also select low PAM matrices such as PAM1.
- For distant related sequences, select low BLOSUM matrices (for example BLOSUM45) or high PAM matrices such as PAM250.
The BLOSUM matrices with low numbers correspond to PAM matrices with high numbers. (See figure 14.17) for correlations between the PAM and BLOSUM matrices. To summarize, if you want to find distant related proteins to a sequence of interest using BLAST, you could benefit of using BLOSUM45 or similar matrices.
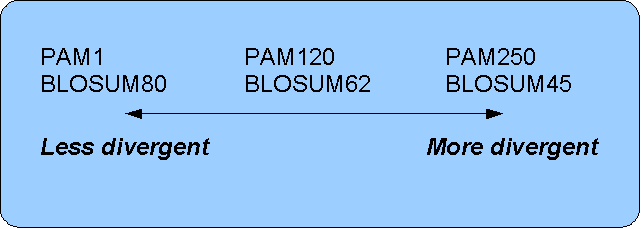
Figure 14.17: Relationship between scoring matrices.
The BLOSUM62 has become a de facto standard scoring matrix
for a wide range of alignment programs. It is the default matrix in BLAST.
Other useful resources
Calculate your own PAM matrix
http://www.bioinformatics.nl/tools/pam.html
BLOKS database
http://blocks.fhcrc.org/
NCBI help site
http://www.ncbi.nlm.nih.gov/Education/BLASTinfo/Scoring2.html
