Significance thresholds
Clicking Next will display the dialog shown in figure 31.11.
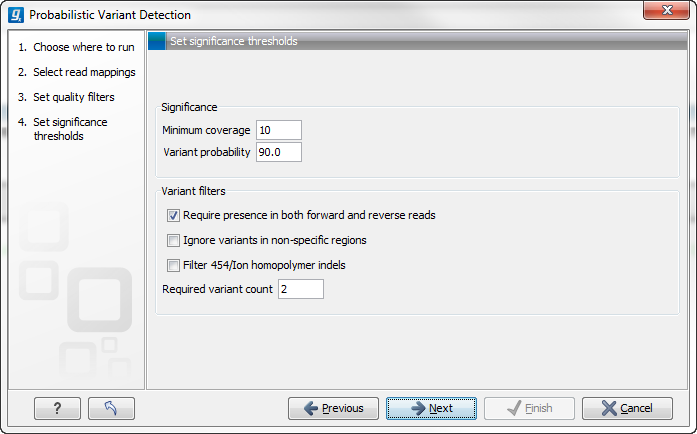
Figure 31.11: Significance thresholds.
The follow parameters can be set:
Significance
- Minimum coverage The minimum number of reads aligned to the site to be considered a potential variant.
- Variant probability The minimum total probability that a variant is different from the reference for that position to be reported.
Variant filters
Below the significance settings, there are filters that can be useful for removing false positives:
- Require presence in both forward and reverse reads. Some systematic sequencing errors can be triggered by a certain combination of bases. This means that sequencing one strand may lead to sequencing errors that are not seen when sequencing the other strand (see [Nguyen et al., 2011] for a recent study with Illumina data). This can easily lead to false positive variant calls, and by checking this filter, the minimum ratio between forward and reverse reads supporting the variant should be at least 0.05. In this way, systematic sequencing errors of this kind can be eliminated. The forward/reverse reads balance is also reported for each variant in the result (see Variant data).
- Ignore variants in non-specific regions. Variants in regions covered by one or more non-specific reads are ignored.
- Filter 454/Ion homopolymer indels. The 454 and Ion Torrent/Proton sequencing platforms exhibit weaknesses when determining the correct number of the same kind of nucleotides in a homopolymer region (e.g. AAA). This leads to a high false positive rate for calling InDels in these regions. This filter is very basic: it removes all indels that are found within or just next to a homopolymer region. A homopolymer region is defined as at least two consecutive identical bases in the reference.
- Required Variant Count. This option is the threshold for the number of reads that display a variant at a given position and is based on absolute counts. If the count required is set to 3, it means that even though the required percentage of the reads has a variant base, it will still not be reported if there are less than 3 reads supporting the variant.
