Export Genotype VCF (beta)
Using Export Genotype VCF (beta) it is possible to export variants in a Genotype track to VCF file.
Below are the main differences between Export Genotype VCF (beta) and the standard VCF exporter in the CLC Workbench:
- Only Genotype tracks can be exported.
- Variant files are exported in VCF 4.3 format.
- Phasing information from the Genotype track is encoded in the VCF.
- The Genotype track locus table (see Locus Table) reflects very well what the contents of the exported VCF will be.
- It is not possible to enforce ploidy.
- Complex variants are represented as described for Reference overlap in standard VCF export: http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Complex_variant_representations_VCF_reference_overlap.html.
To export variants in a Genotype track to VCF, click Export in the Toolbar and choose the tool Export Genotype VCF (beta). Specify the Genotype tracks that should be exported and click Next. This will open a dialog as shown in figure 10.10.
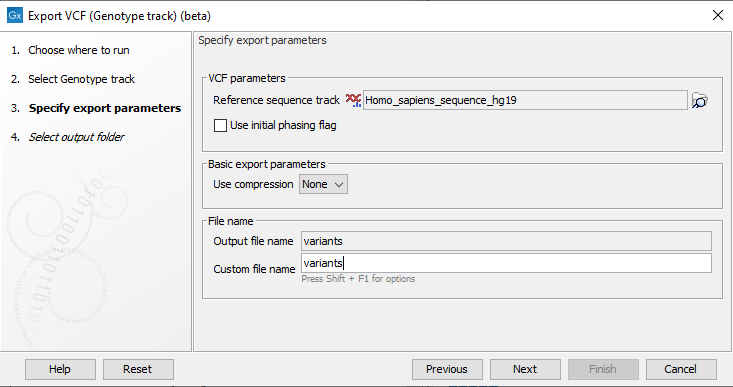
Figure 10.10: Setting parameters for export.
The following parameters can be adjusted in the dialog:
- Reference sequence track: Since the VCF format specifies that reference and allele sequences cannot be empty, deletions and insertions have to be padded with bases from the reference sequence. The export needs access to the reference sequence track in order to find the neighboring bases.
- Use Initial phasing flag: Enable this option to specify phasing status for the first allele in haploid and mixed phasing genotypes by using an initial phasing flag (e.g. GT=|1 and GT=/0|1|2). Disable to increase compatibility with external applications at the cost of lost phasing information. When disabled, some phased alleles in mixed phasing genotypes are specified as unphased, and phased haploid genotypes are specified using a missing allele (e.g. GT=1|. and GT=0/1|2).
