Merge Immune Repertoire
The Merge Immune Repertoire tool can be used for reducing false positives due to sequencing errors and variability in read quality and length:
- Read quality and length variability can impact the identified segments.
- Reference segments may have a large degree of sequence identity due to recent duplication events [Glusman et al., 2001]. In order to uniquely identify a segment, reads need to be sufficiently long to cover the regions where paralogue segments differ. Shorter reads may lead to clonotypes containing multiple (ambiguous) segments.
- Identification of the C segment requires reads that are sufficiently long. Shorter reads will be reported without an identified C segment.
- Sequencing errors in the CDR3 region can lead a highly expressed clonotype to be reported as multiple clonotypes.
Merge Immune Repertoire is available from the Tools menu at:
Tools | Biomedical Genomics Analysis (![]() ) | Immune Repertoire Analysis (
) | Immune Repertoire Analysis (![]() ) | Merge Immune Repertoire (
) | Merge Immune Repertoire (![]() )
)
This opens a dialog where a TCR clonotypes (![]() ) or BCR clonotypes (
) or BCR clonotypes (![]() ) element can be selected. The following options can be configured (figure 7.8):
) element can be selected. The following options can be configured (figure 7.8):
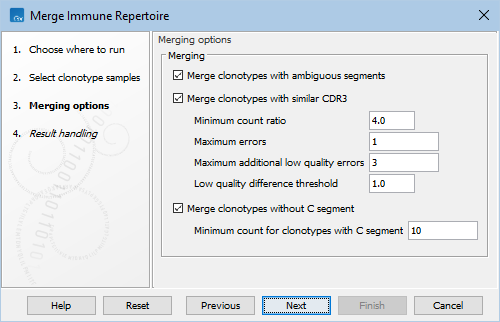
Figure 7.8: Options for Merge Immune Repertoire.
- Merge clonotypes with ambiguous segments. If selected, merge clonotypes with compatible V, J and C segments and the same CDR3 nucleotide sequence, where one clonotype has a unique segment and the other has ambiguous segments that include the former clonotype's segment.
If two clonotypes are merged, the unique segment is preserved in the merged clonotype, regardless of the counts of the two clonotypes.
Note that using this option can lead to some reads not being included in the alignments view. See Alignments after merging for details.
- Merge clonotypes with similar CDR3. If selected, merge clonotypes with the same identified V, J, and C segments and with similar CDR3 nucleotide sequences. As the D segment is found within the CDR3, clonotypes are not required to have the same identified D segment.
If two clonotypes are merged, the CDR3 sequence and identified D segment of the larger clonotype are preserved in the merged clonotype.
- Minimum count ratio. A smaller clonotype is merged into a larger clonotype if the count of the larger clonotype is at least this number of times larger than the count of the smaller clonotype.
E.g. if the minimum count ratio is 4 and a clonotype has count 8, only clonotypes with a count of at most 2 (8 / 4 = 2) will be considered for merging.
- Maximum errors. Two clonotypes will be considered for merging if there are at most this many differences between their CDR3 sequences.
- Maximum additional low quality errors. Two clonotypes where the number of differences between their CDR3 sequences exceeds Maximum errors can still be considered for merging, if the number of additional errors at positions with low quality in the smaller clonotype does not exceed this number.
- Low quality difference threshold. A position is considered of low quality if the average quality is more than this number of standard deviations lower than the average quality at each position in the CDR3 sequence.
Note that Maximum additional low quality errors and Low quality difference threshold have no effect if the CDR3 quality scores are not available, see Table for Clonotypes.
- Minimum count ratio. A smaller clonotype is merged into a larger clonotype if the count of the larger clonotype is at least this number of times larger than the count of the smaller clonotype.
- Merge clonotypes without C segment. If selected, merge clonotypes with the same identified V, J, and D segments and the same CDR3 nucleotide sequences, where one clonotype has an identified C segment and the other one does not.
If two clonotypes are merged, the identified C segment is preserved in the merged clonotype.
- Minimum count for clonotypes with C segment.
A smaller clonotype with a C segment is merged with a larger clonotype without a C segment if the count of the smaller clonotype is at least this number.
Note that a smaller clonotype without a C segment is always merged with a larger clonotype with a C segment.
- Minimum count for clonotypes with C segment.
A smaller clonotype with a C segment is merged with a larger clonotype without a C segment if the count of the smaller clonotype is at least this number.
Subsections
