Immune Repertoire Analysis
Using RNA-Seq data as input, the Immune Repertoire Analysis tool can be used to characterize either the T or B cell receptor repertoire.
The tool requires a reference data sequence list (![]() ) containing reference sequences for the V, D, J and C segments.
) containing reference sequences for the V, D, J and C segments.
Whether the tool identifies T or B cell receptors depends on the types of reference segments present in the provided sequence list. The tool does not accept sequence lists containing reference sequences for both TCR and BCR.
The Reference Data Manager (see Reference Data Management) offers two QIAGEN sets for this tool. Each set contains a sequences list for Immune Repertoire Analysis:
- QIAseq Immune Repertoire Analysis for analysis of TCR human data.
- QIAseq Immune Repertoire Analysis Mouse for analysis of TCR mouse data.
If reference data is needed for BCR or for a different species than those above, Import Immune Reference Segments can be used to import reference data, see Import Immune Reference Segments.
|
The tool assumes that one read spans all segment types (V, D, J and C) in order to successfully report the clonotype. It is therefore recommended to collapse overlapping paired-end reads using Merge Overlapping Pairs. |
Identification of clonotypes
Clonotyping a read consists of identifying which V, D, J and C segments from the reference data are used and extracting the CDR3 sequence found between the conserved amino acids.
V and C segments are rather long (![]() bp), whereas J segments are relatively short (
bp), whereas J segments are relatively short (
![]() bp) and D segments are even shorter (
bp) and D segments are even shorter (
![]() bp).
The segments identification is therefore performed using different strategies.
bp).
The segments identification is therefore performed using different strategies.
First, the tool identifies the V and J segments. These segments are required for successfully clonotyping a read, because otherwise the CDR3 cannot be determined.
For V segments, the Map Reads to Reference tool is used internally.
For the J segment, a strategy similar to IMSEQ [Kuchenbecker et al., 2015] is used. First, a pairwise alignment with a 15 bp subsequence of the full segment called a Segment Core Fragment (SCF) is performed to find candidates for full pairwise alignments. If the pairwise alignment of an SCF to the read has a sufficiently small number of errors, it is nominated as a candidate. A full pairwise alignment is then made for all the segments corresponding to the candidate SCFs. If there is a sufficiently good match among the full alignments it will be assigned to the read.
Once both V and J segments are identified, only valid matches are kept:
- the V and J segments are for the same chain;
- the J segment is located on the read after the V segment.
The D and C segments are then identified for the reads with assigned V and J segments. These segments are optional.
For D segments, a local alignment is performed between the region of the read found between V and J, and the reference D segments for the same chain.
For C segments, the Map Reads to Reference tool is used internally. As the C segment is long and not variable, matches for the C segment for chains other than that identified for V and J indicate a false positive and the read is hence discarded.
Read length variability and sequencing errors can lead to one clonotype being reported as two separate clonotypes. See Merge Immune Repertoire for details on how to merge such clonotypes into one single clonotype.
Running the tool
Immune Repertoire Analysis is available from the Tools menu at:
Tools | Biomedical Genomics Analysis (![]() ) | Immune Repertoire Analysis (
) | Immune Repertoire Analysis (![]() ) | Immune Repertoire Analysis (
) | Immune Repertoire Analysis (![]() )
)
This opens a dialog where the reads can be selected. The following options can be configured (figure 7.6):
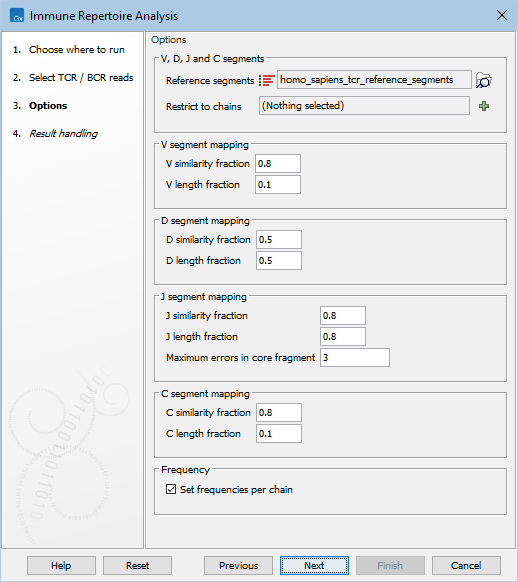
Figure 7.6: Options for Immune Repertoire Analysis.
- Reference segments. A sequence list containing the V, D, J and C segments, either from the reference data or imported using Import Immune Reference Segments.
- V / D / J / C similarity fraction. Minimum identity fraction between the aligned region of the read and the segment.
- V / D / J / C length fraction. Minimum fraction of the segment that must match the read.
- Maximum errors in core fragment.
Maximum number of errors allowed in the Segment Core Fragment (SCF) used for finding segment candidates for full pairwise alignment.
- Set frequencies per chain. Clonotype frequencies (see Table For Clonotypes) are calculated such that they add up to 100% across all chains. If Set frequencies per chain is ticked, the frequencies are instead calculated such that they add up to 100% for each individual chain.
|
The optimal values for the Similarity fraction and Length fraction are different for the different segment types.
As the V and C segments are at the ends of the read, they might not be covered entirely and the length fraction is expected to be considerably smaller than 1. On the other hand, the length fraction would typically be close to 1 for J. For V, J and C, the similarity fraction is usually close to 1 as not a lot of mutations are expected in these segments.
As the D segment is located in a region of high variability, both the similarity and length fractions would typically be lower to account for the high mutation rate. |
Subsections
