Merging of clonotypes
The three merging options can be used together.
The following is an example of using both Merge clonotypes with similar CDR3 and Merge clonotypes without C segment. Let us consider two clonotypes with the same identified V and J segments, sufficiently similar CDR3 nucleotide sequences, and where only one has an identified C segment:
- If both options are selected, the clonotypes will be merged.
- If only Merge clonotypes with similar CDR3 is selected, the clonotypes will not be merged since they have different identified segments.
- If only Merge clonotypes without C segment is selected, the clonotypes will not be merged since they have different CDR3 sequences.
Multiple clonotypes with similar CDR3 sequences can be merged in different ways, depending on the values set for the different options. Consider the example from figure 7.9:
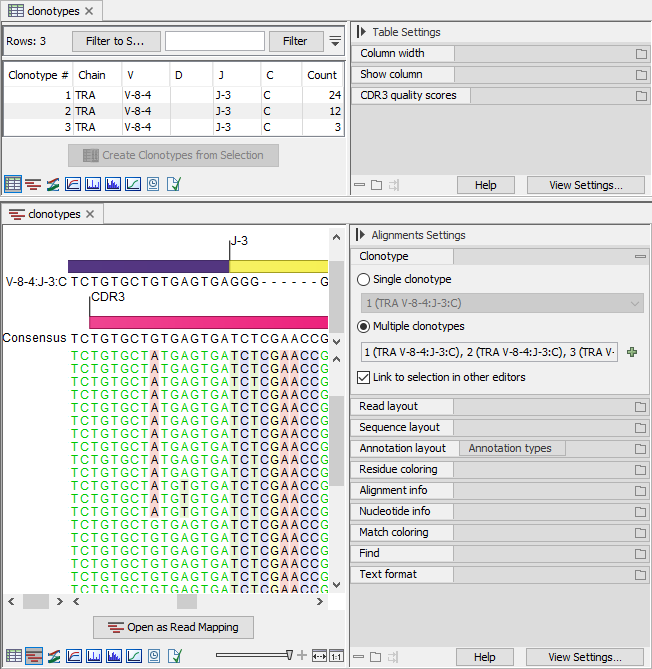
Figure 7.9: Three clonotypes sharing the identified segments and with similar CDR3 sequences. Top: Clonotypes table view. Bottom: Clonotypes alignment view for all three clonotypes, highlighting the differences in the CDR3 sequence.
- The CDR3 sequences of clonotypes 1 and 2 differ by one nucleotide.
- The CDR3 sequences of clonotypes 2 and 3 differ by one nucleotide at a different position, such that the CDR3 sequences of clonotypes 1 and 3 differ by two nucleotides.
- The counts for clonotypes 1, 2 and 3 are 24, 12 and 3, respectively.
Consecutive merges
Clonotypes merging is performed consecutively and hence multiple clonotypes may be merged into one. Setting Minimum count ratio to 1.5 and Maximum errors to 1:
- Clonotype 3 is merged into clonotype 2, leading to clonotype 2 having a count of 15.
- Clonotype 2 is merged into clonotype 1, because 24 / 15 > 1.5. Since clonotype 3 has already been merged into clonotype 2, this effectively also merges clonotype 3 into clonotype 1, even though the number of differences between their CDR3 sequences (2) exceeds Maximum errors.
Merges into multiple clonotypes
A clonotype may be merged into multiple clonotypes. Setting Minimum count ratio to 4 and Maximum errors to 2, clonotype 3 is merged into both clonotypes 1 and 2, and its count is distributed between clonotypes 1 and 2, proportional to their respective counts:
- Clonotype 2 receives 1 (3 x 12 / (12 + 24)), for a total count of 13.
- Clonogype 1 receives 2 (3 x 24 / (12 + 24)), for a total count of 26.
Clonotype 2 is not merged into clonotype 1 because 26 / 13 < 4.
Identical due to preceding merging
Two originally different clonotypes can end up being identical due to preceding merging, and then they will be merged into one. Consider the example from figure 7.10:
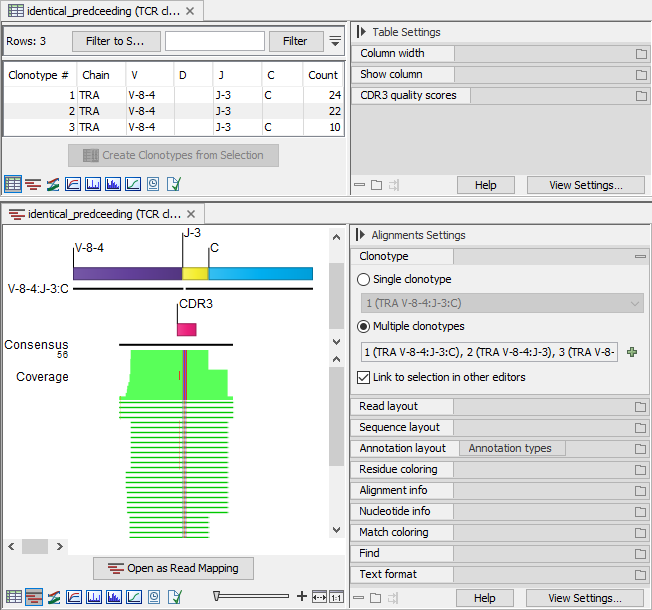
Figure 7.10: Three clonotypes sharing the identified segments, with similar CDR3 sequences, and where one is missing the C segment. Top: Clonotypes table view. Bottom: Clonotypes alignment view for all three clonotypes, highlighting the differences in the CDR3 sequence and the shorter reads not covering the C segment.
- Clonotypes 1 and 2 have the same CDR3 sequence, but only clonotype 1 has a C segment.
- Clonotype 3 has a CDR3 sequence that is sufficiently similar to that of clonotypes 1 and 2, and the same C segment as clonotype 1.
- The counts for clonotypes 1, 2 and 3 are 24, 22 and 10, respectively.
Running the tool with both Merge clonotypes with similar CDR3 and Merge clonotypes without C segment, setting Minimum count ratio to 2, Maximum errors to 1 and Minimum count for clonotypes with C segment to 10:
- Clonotype 3 is merged into:
- Clonotype 1 due to similar CDR3.
- Clonotype 2 due to similar CDR3 and missing C segment.
- Clonotype 2 now has a C segment and is identical to clonotype 1 due to the preceding merging. The clonotypes are therefore merged.
