Output from Import Immune Reference Segments
The importer outputs a sequence list that can be used for immune repertoire analysis. These can be added to a Custom Set for use in workflows.
The sequence list contains the reference sequences for the V, D, J and C segments, named in the format <chain>-<type>-<ID>*<allele>, for example "TRA-V-1*01". Note that for B cells constant genes, the letter corresponding to the encoded isotype is used instead of the segment type.
If the gene segment does not have an allele or Import only the first allele is ticked, *<allele> is not added to the name.
By ticking "Show annotations" and "Region" in the Side Panel "Annotation layout" and "Annotation types" palettes, respectively, the location of the conserved amino acid can be visualized (figure 7.4).
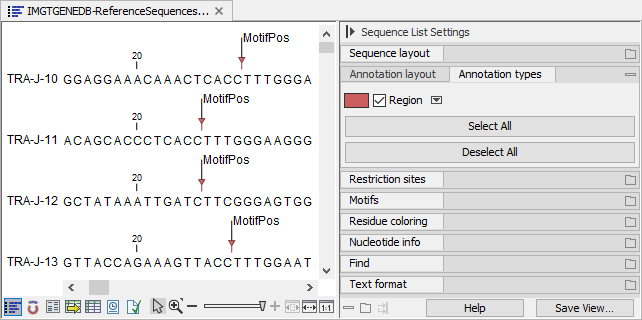
Figure 7.4: Visualizing the location of the conserved amino acid.
The table view (![]() ) of the sequence list contains the chain, segment type and allele number for each gene segment. For the IMGT format, it also contains the accession number(s), species and functionality (figure 7.5). These can be used to filter the gene segments to a subset of interest.
) of the sequence list contains the chain, segment type and allele number for each gene segment. For the IMGT format, it also contains the accession number(s), species and functionality (figure 7.5). These can be used to filter the gene segments to a subset of interest.
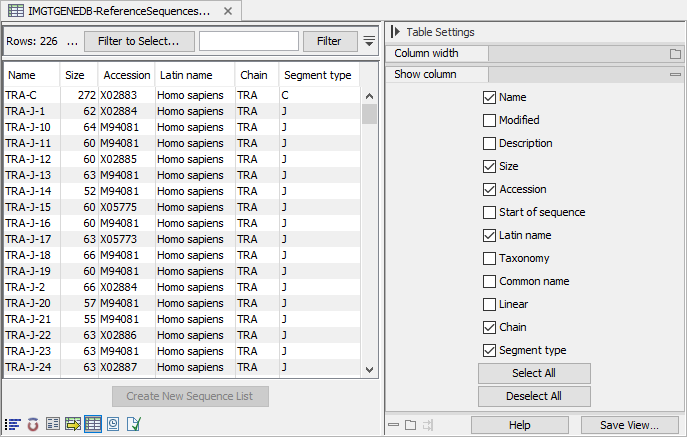
Figure 7.5: Table view of an imported sequence list using the IMGT format. Note that not all columns are shown.
