Perform QIAseq Targeted TCR Analysis
The Perform QIAseq Targeted TCR Analysis workflow can be used to characterize the T cell receptor (TCR) immune repertoire for RNA-Seq data produced with the QIAseq Targeted RNA-seq Panel for T-cell Receptor.
The workflow includes all necessary steps for processing the RNA-Seq reads and characterizing the repertoire:
- UMIs are removed using Remove and Annotate with Unique Molecular Index
- Common sequence is removed using Trim Reads, see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Trim_Reads.html
- UMI reads are created using Create UMI Reads from Reads
- Overlapping paired UMI reads are merged using Merge Overlapping Pairs, see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Merge_Overlapping_Pairs.html
- Both merged and not merged UMI reads are trimmed using Trim Reads, see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Trim_Reads.html
- Clonotypes are identified using Immune Repertoire Analysis
- Identified clonotypes are merged using Merge Immune Repertoire
- Merged clonotypes are filtered to remove false positives using Filter Immune Repertoire
- A summary report is created using Create Sample Report.
Launching the workflow
The Perform QIAseq Targeted TCR Analysis template workflow is available under the Workflows menu at:
Workflows | Template Workflows | Biomedical Workflows (![]() ) | QIAseq Sample Analysis (
) | QIAseq Sample Analysis (![]() ) | Other QIAseq Workflows (
) | Other QIAseq Workflows (![]() ) | Perform QIAseq Targeted TCR Analysis (
) | Perform QIAseq Targeted TCR Analysis (![]() )
)
For general information about launching workflows, see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Launching_workflows_individually_in_batches.html
Options can be configured in the following dialogs:
- Choose where to run. If you are connected to a CLC Server via the CLC Workbench, you will be asked where you would like to run the analysis. We recommend that you run the analysis on a CLC Server when possible.
- Select Reads. Select the RNA-Seq reads. When analyzing more than one sample at a time, check the Batch checkbox in the lower left corner of the dialog.
- Specify reference data handling.
Select the relevant Reference Data Set. The Reference Data Manager (see Reference Data Management) offers two QIAGEN sets:
- QIAseq Immune Repertoire Analysis for analysis of TCR human data.
- QIAseq Immune Repertoire Analysis Mouse for analysis of TCR mouse data.
- Configure batching. If running the workflow in Batch mode, you will be asked to define the batch units.
- Batch overview, if running in batch mode. Verify that the batching is as intended.
- Create UMI Reads from Reads. Adjust Minimum UMI group size if needed. UMI groups supported by fewer reads than this number will be discarded. See Create UMI Reads from Reads for details.
- Immune Repertoire Analysis. Set Restrict to chains if only specific chains have been sequenced. See Immune Repertoire Analysis for more details.
- Filter Immune Repertoire. Uncheck Use minimum count if the identified clonotypes should not be filtered. Otherwise, adjust Minimum count if needed. Clonotypes supported by fewer UMI reads than this number will be discarded. See Filter Immune Repertoire for details.
- Create Sample Report. Select relevant summary items and specify thresholds for quality control. Summary items, thresholds, and an indication of whether specified thresholds were met, will be shown in the quality control section of the sample report. The default summary items are appropriate for many data sets, but may need to be adjusted.
- Result handling. Choose if a workflow result metadata and/or log should be saved.
- Save location for new elements. Choose where to save the data, and press Finish to start the analysis.
Launching using the QIAseq Panel Analysis Assistant
The workflow is also available in the QIAseq Panel Analysis Assistant under Immune.
When launching from the assistant, the analysis can be restricted to the sequenced chains (figure 14.1).
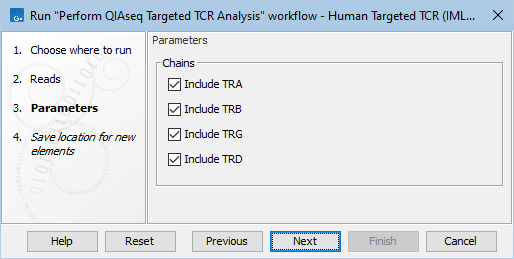
Figure 14.1: Selecting the chains to be used for the analysis when executing the workflow from the QIAseq Panel Analysis Assistant.
Subsections
