Upload to IPA
QIAGEN Ingenuity Pathway Analysis (IPA) aids the interpretation of differential expression results by identifying enriched pathways and determining the activity of upstream regulators. It assesses potential impacts on downstream diseases, biological functions, and phenotypes.
Upload to IPA takes statistical comparison tables (![]() ) or tracks (
) or tracks (![]() ) as input and uploads them to IPA.
) as input and uploads them to IPA.
To run the tool, go to:
Tools | Biomedical Genomics Analysis (![]() ) | IPA and QCI Interpret Upload (
) | IPA and QCI Interpret Upload (![]() ) | Upload to IPA (
) | Upload to IPA (![]() )
)
After selecting the inputs, use the IPA login wizard to log in to IPA. Select the relevant IPA server (US or China) and click the Log in button. This opens an IPA login page in an external browser. Once authenticated, the following options can be configured in the Configuration wizard (figure 9.1):
- Project Name The name of the project in IPA. The placeholder {1} will be substituted with the current date in the format YYYY-MM-DD.
- Reference data The reference data can be set to:
- Ingenuity Knowledge Base (Genes only) The Ingenuity Knowledge Base is used as reference data. This is only suitable for inputs pertaining to gene expression. See Upload using the Ingenuity Knowledge Base for details.
- Uploaded dataset The dataset itself is used as reference data.
- Upload only Only upload the dataset to IPA.
- Upload and analyze Upload to IPA and perform pathway analysis on features meeting the filter criteria specified using the following options:
- Minimum mean expression value Only features with at least this "Max group mean" are analyzed.
- Maximum p-value Only features with at most this p-value are analyzed. The P-value type can be set to P-value, FDR p-value, or Bonferroni.
- Only features with an absolute fold change above a certain threshold are analyzed. The Fold change type can be set to Fold change or Log
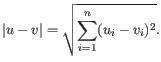 fold change.
fold change.
- Automatically calculate fold change threshold If checked, the threshold is calculated for each input separately, such that the number of analyzed features is as close as possible to the Target number of features.
- Fold change threshold The provided threshold is used.
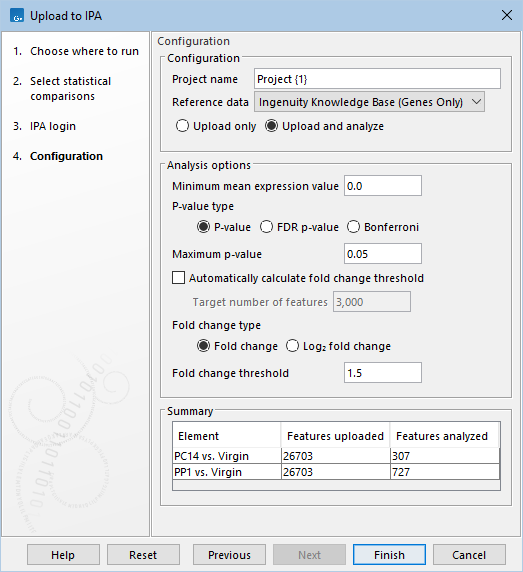
Figure 9.1: Configuration options for Upload to IPA.
Note that all features in the input are uploaded. To limit statistical comparisons to only the differentially expressed features, use Filter on Custom Criteria.
The summary at the bottom of the wizard (figure 9.1) provides an overview of the number of features to be uploaded, and, if applicable, the number of features to be analyzed and the automatically calculated fold change threshold.
Subsections
