Sequence region types
The various annotations on sequences
cover parts of the sequence. Some cover an interval, some cover
intervals with unknown endpoints, some cover more than one interval
etc. In the following, all of these will be referred to as
regions. Regions are generally illustrated by markings (often
arrows) on the sequences. An arrow pointing to the right indicates
that the corresponding region is located on the positive strand of
the sequence. Figure 9.2 is an example of three
regions with separate colors.
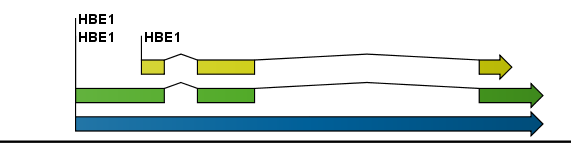
Figure 9.2: Three regions on a human beta globin DNA sequence (HUMHBB).
Figure 9.3 shows an artificial sequence with all the different kinds of regions.

Figure 9.3: Region #1: A single residue, Region #2: A range of residues including both endpoints, Region #3: A range of residues starting somewhere before 30 and continuing up to and including 40, Region #4: A single residue somewhere between 50 and 60 inclusive, Region #5: A range of residues beginning somewhere between 70 and 80 inclusive and ending at 90 inclusive, Region #6: A range of residues beginning somewhere between 100 and 110 inclusive and ending somewhere between 120 and 130 inclusive, Region #7: A site between residues 140 and 141, Region #8: A site between two residues somewhere between 150 and 160 inclusive, Region #9: A region that covers ranges from 170 to 180 inclusive and 190 to 200 inclusive, Region #10: A region on negative strand that covers ranges from 210 to 220 inclusive, Region #11: A region on negative strand that covers ranges from 230 to 240 inclusive and 250 to 260 inclusive.
Note When editing annotated nucleotide sequences, the annotation content is not updated automatically (but its position is). Please refer to Edit annotations for details on annotation editing. Before exporting annotated nucleotide sequences in GenBank format, ensure that the annotations in the Annotations Table reflect the edits that have been made to the sequence.
