Different types of signal peptides
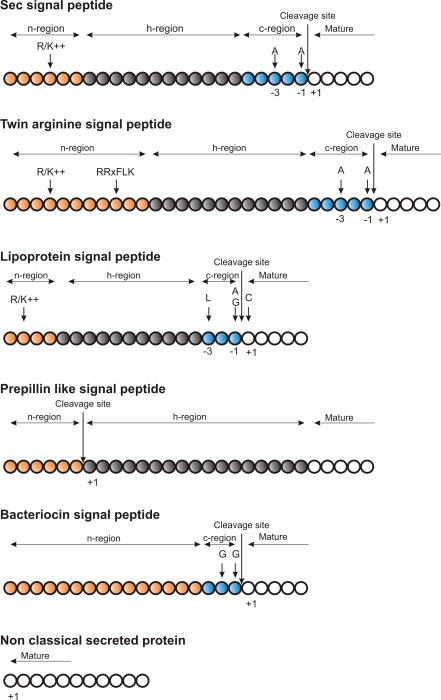
Figure 15.3: Schematic representation of various signal
peptides. Red color indicates n-region, gray color indicates
h-region, cyan indicates c-region. All white circles are part of the
mature protein. tex2html_wrap_inline$+1$ indicates the first position of the mature
protein. The length of the signal peptides is not drawn to scale.
Soon after Günter Blobel's initial discovery of signal peptides, more targeting signals were found. Most cell types and organisms employ several ways of targeting proteins to the extracellular environment or subcellular locations. Most of the proteins targeted for the extracellular space or subcellular locations carry specific sequence motifs (signal peptides) characterizing the type of secretion/targeting it undergoes.
Several new different signal peptides or targeting signals have been found during the later years, and papers often describe a small amino acid motif required for secretion of that particular protein. In most of the latter cases, the identified sequence motif is only found in this particular protein and as such cannot be described as a new group of signal peptides.
Describing the various types of signal peptides is beyond the scope of this text but several review papers on this topic can be found on PubMed. Targeting motifs can either be removed from, or retained in the mature protein after the protein has reached the correct and final destination. Some of the best characterized signal peptides are depicted in figure 15.3.
Numerous methods for prediction of protein targeting and signal peptides have been developed; some of them are mentioned and cited in the introduction of the SignalP research paper [Bendtsen et al., 2004b]. However, no prediction method will be able to cover all the different types of signal peptides. Most methods predicts classical signal peptides targeting to the general secretory pathway in bacteria or classical secretory pathway in eukaryotes. Furthermore, a few methods for prediction of non-classically secreted proteins have emerged [Bendtsen et al., 2004a,Bendtsen et al., 2005].
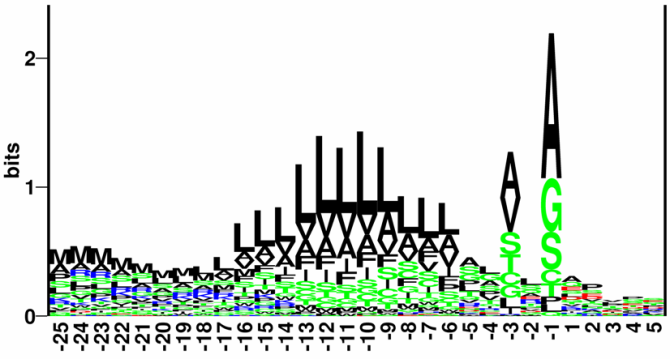
Figure 15.4: Sequence logo of eukaryotic signal peptides, showing conservation of amino acids in bits
[Schneider and Stephens, 1990]. Polar and hydrophobic residues are shown in
green and black, respectively, while blue indicates positively
charged residues and red negatively charged residues. The logo is
based on an ungapped sequence alignment fixed at the -1 position of
the signal peptides.
