Selecting sequences for
evaluation
The
evaluation is started from the Toolbox:
Toolbox | Classical Sequence Analysis (![]() ) | RNA Structure (
) | RNA Structure (![]() )|
Evaluate Structure Hypothesis (
)|
Evaluate Structure Hypothesis (![]() )
)
This opens the dialog shown in figure 21.22.

Figure 21.22: Selecting RNA or DNA sequences for evaluating structure hypothesis.
If you have selected sequences before choosing the Toolbox action, they are now listed in the Selected Elements window of the dialog. Use the arrows to add or remove sequences or sequence lists from the selected elements. Note, that the selected sequences must contain a structure hypothesis in the form of manually added constraint annotations.
Click Next to adjust evaluation parameters (see figure 21.23).
The partition function algorithm includes a number of advanced options:
- Avoid isolated base pairs. The algorithm filters out isolated base pairs (i.e. stems of length 1).
- Apply different energy rules for Grossly Asymmetric Interior Loops (GAIL). Compute the minimum free energy applying different rules for Grossly Asymmetry Interior Loops (GAIL).
A Grossly Asymmetry Interior Loop (GAIL) is an interior loop that is
 or
or 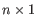 where
where 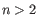 (see
http://mfold.rna.albany.edu/doc/mfold-manual/node5.php)
(see
http://mfold.rna.albany.edu/doc/mfold-manual/node5.php)
- Include coaxial stacking energy rules. Include free energy increments of coaxial stacking for adjacent helices [Mathews et al., 2004].

Figure 21.23: Adjusting parameters for hypothesis evaluation.
