De novo assembly parameters
To start the assembly:
Toolbox | De Novo Sequencing (![]() ) | De Novo Assembly (
) | De Novo Assembly (![]() )
)
In this dialog, you can select one or more sequence lists or single sequences.
Click Next to set the parameters for the assembly. This will show a dialog similar to the one in figure 27.20.
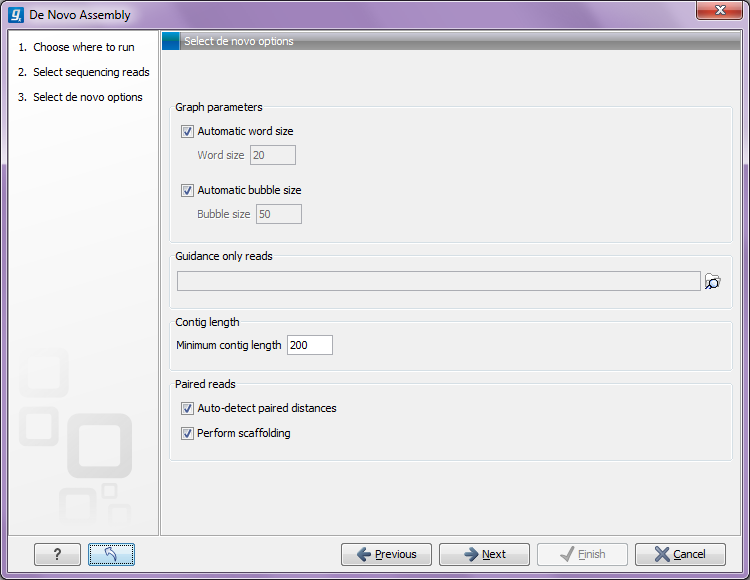
Figure 27.20: Setting parameters for the assembly.
At the top, you select the Word size and the Bubble size to be used. The principles of setting the word size are described in How it works. When using automatic calculation, you can see the word size in the History (![]() ) of the result files. Please note that the range of word sizes is 12-24 on 32-bit computers and 12-64 on 64-bit computers.
) of the result files. Please note that the range of word sizes is 12-24 on 32-bit computers and 12-64 on 64-bit computers.
The meaning of the bubble size parameter is explained in Bubble resolution. The automatic bubble size is set to 50, unless one of the following conditions apply:
- some of the reads are from either 454, Ion torrent or PacBio;
- the reads are not all Sanger reads and average read length of all input reads is >160bp.
In these cases the bubble size is set to the average read length of all input reads. The value used is also recorded in the History (![]() ) of the result files.
) of the result files.
The next option is to specify Guidance only reads. The reads supplied here will not be used to create the de Bruijn graph and subsequent contig sequence but only used to resolved ambiguities in the graph (see Resolve repeats using reads and Optimization of the graph using paired reads). With mixed data sets from different sequencing platforms, we recommend using sequencing data with low error rates as the main input for the assembly, whereas data with more errors should be specified only as Guidance only reads. This would typically be long reads or paired data sets.
You can also specify the Minimum contig length when doing de novo assembly. Contigs below this length will not be reported. The default value is 200 bp. For very large assemblies, the number of contigs can be huge (over a million), in which case the data structures when mapping reads back to contigs will be very large and take a very long time to handle. In this case, it is a great advantage to raise the minimum contig length to reduce the number of contigs that have to be incorporated into this data structure.
At the bottom, there is an option to Perform scaffolding. The scaffolding step is explained in greater detail in Optimization of the graph using paired reads. This will also cause scaffolding annotations to be added to the contig sequences (except when you also choose to Update contigs, see below).
Finally, there is an option to Auto-detect paired distances. This will determine the paired distance (insert size) of paired data sets. If several paired sequence lists are used as input, a separate calculation is done for each one to allow for different libraries in the same run. The History (![]() ) view of the result will list the distance used for each data set.
) view of the result will list the distance used for each data set.
If the automatic detection of pairs is not checked, the assembler will use the information about minimum and maximum distance recorded on the input sequence lists (see General notes on handling paired data).
For mate-pair data sets with large insert sizes, it may not be possible to infer the correct paired distance. In this case, the automatic distance calculation should not be used.
The best way of checking this is to run a read mapping using the contigs from the de novo assembly as reference and the mate-pair library as reads, and then check the Detailed mapping report. There is a paired distance distribution graph that can be used to check whether the distance estimated by the assembler fits in the distribution found in the read mapping.
When you click Next, you will see the dialog shown in figure 27.21
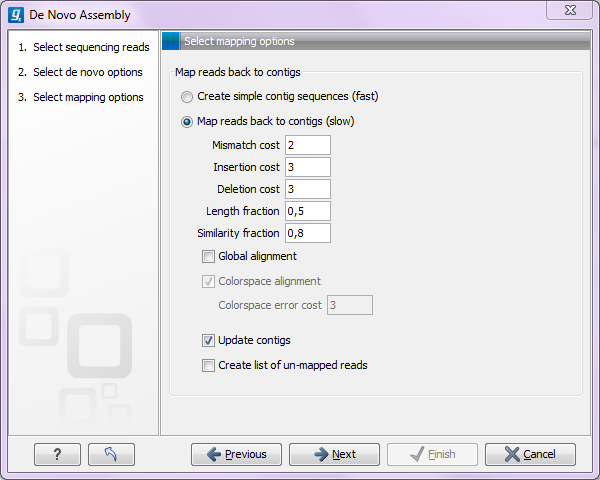
Figure 27.21: Parameters for mapping reads back to the contigs.
At the top, you choose whether a read mapping should be performed after the initial contig creation. If chosen, you can specify some parameters for the read mapping. The mapping options offered in the wizard are explained in The Map Reads to Reference tool manual pages. The placement of reads that map in more than one position equally well are placed randomly (see section about Non-Specific Matches) and the type of gap costs used here are linear.
At the bottom, you can choose to Update contigs based on the subsequent mapping of the input reads back to the contigs generated by the de novo assembly. In general terms, this has the effect of updating the contig sequences based on the evidence provided by the subsequent mapping back of the read data to the de novo assembled contigs. The following are the impacts of choosing this option:
- Contig regions must be supported by at least one read mapping back to them in order to be included in the output. If more than half of the reads in a column of the mapping contain a gap, then a gap will be entered into the contig sequence. Contig regions where no reads map will be removed. Note that if such a region occurs within a contig, it is removed and the surrounding regions are joined together.
- The most common nucleotide among the mapped reads at a given position is the one assigned to the contig sequence. In NGS data, it would be very unlikely that at a given position there would be an equal number of reads with different nucleotides. Should this occur however, then the nucleotide that comes first in the alphabet would be included in the consensus.
Note that if this option is selected, the contig lengths may get below the threshold specified in figure 27.20 because this threshold is applied to the original contig sequences. If the Update contigs based on mapped reads option is not selected, the original contig sequences from the assembler will be preserved completely also in situations where the reads that are mapped back do not support the contig sequences.
