Run the Identify Variants (WES) workflow
- To run the Identify Variants (WES) template workflow, go to:
Workflows |Template Workflows | Biomedical Workflows (
 ) | Whole Exome Sequencing (
) | Whole Exome Sequencing ( ) | Somatic Cancer (
) | Somatic Cancer ( ) | Identify Variants (WES) (
) | Identify Variants (WES) ( )
)
- Select the sequencing reads from the sample that should be analyzed (figure 18.23).
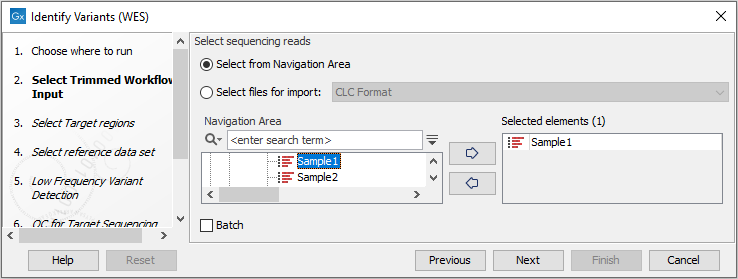
Figure 18.23: Please select all sequencing reads from the sample to be analyzed.If several samples should be analyzed, the tool has to be run in batch mode. This is done by checking "Batch" and selecting the folder that holds the data you wish to analyze.
- Next, in the Target regions dialog you need to specify the target regions for your application. The variant calling will be restricted to these regions (figure 18.24).
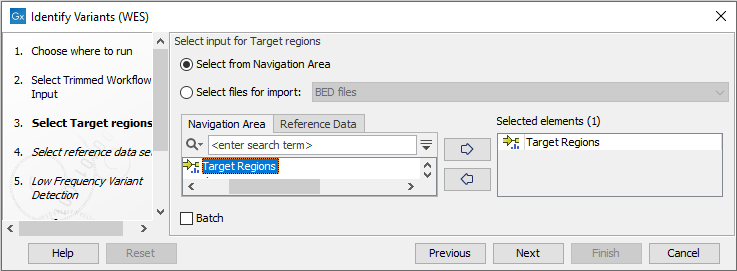
Figure 18.24: Select the track with the targeted regions from your experiment. - In the next dialog, you have to select which reference data set should be used to identify variants (figure 18.25).
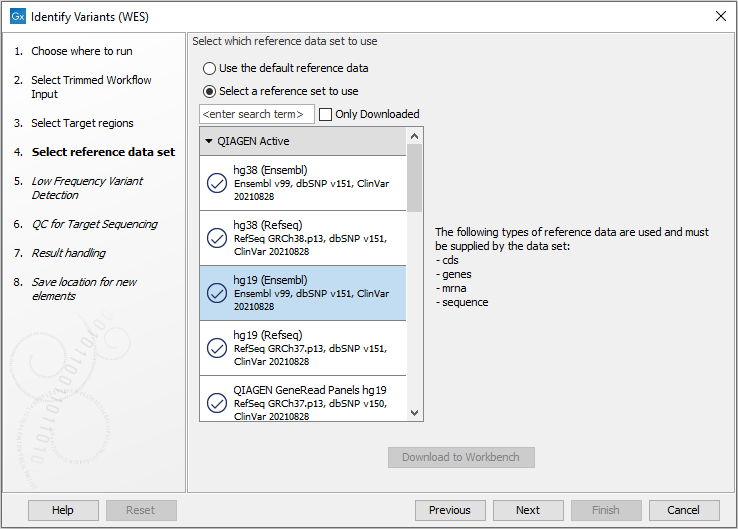
Figure 18.25: Choose the relevant reference Data Set to identify variants in your sample. - In the next wizard step (figure 18.26), you can specify the parameters for variant detection.
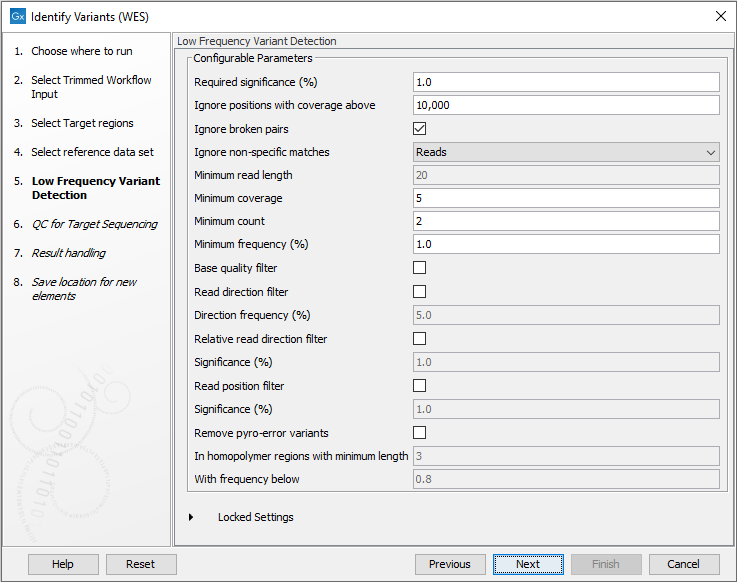
Figure 18.26: Specify the parameters for variant detection. - In the QC for Target Sequencing step (figure 18.27) you can specify the minimum read coverage, which should be present in the targeted regions.
Figure 18.27: Specify the minimum coverage for the QC for Targeted sequencing. - In the last wizard step you can check the selected settings by clicking on the button labeled Preview All Parameters.
In the Preview All Parameters wizard you can only check the settings, and if you wish to make changes you have to use the Previous button from the wizard to edit parameters in the relevant windows.
- Choose to Save your results and click Finish.
