Run the Annotate Variants (WES) workflow
- To run the Annotate Variants (WES) template workflow, go to:
Workflows | Template Workflows | Biomedical Workflows (
 ) | Whole Exome Sequencing (
) | Whole Exome Sequencing ( ) | General Workflows (WES) (
) | General Workflows (WES) ( ) | Annotate Variants (WES) (
) | Annotate Variants (WES) ( )
)
- In the first wizard step, select the input variant track, annotation track, expression track or statistical comparison track to annotate (figure 18.1).
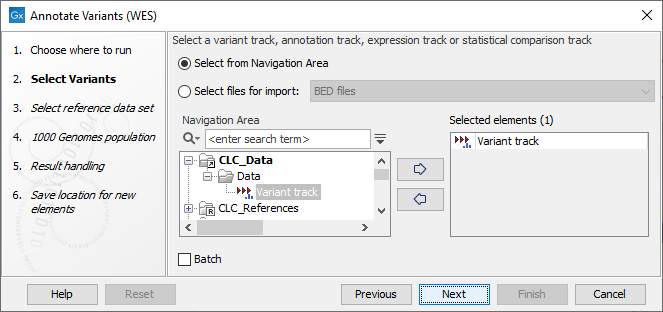
Figure 18.1: Select the relevant track to annotate. - In the next dialog, you have to select which data set should be used to annotate variants (figure 18.2).
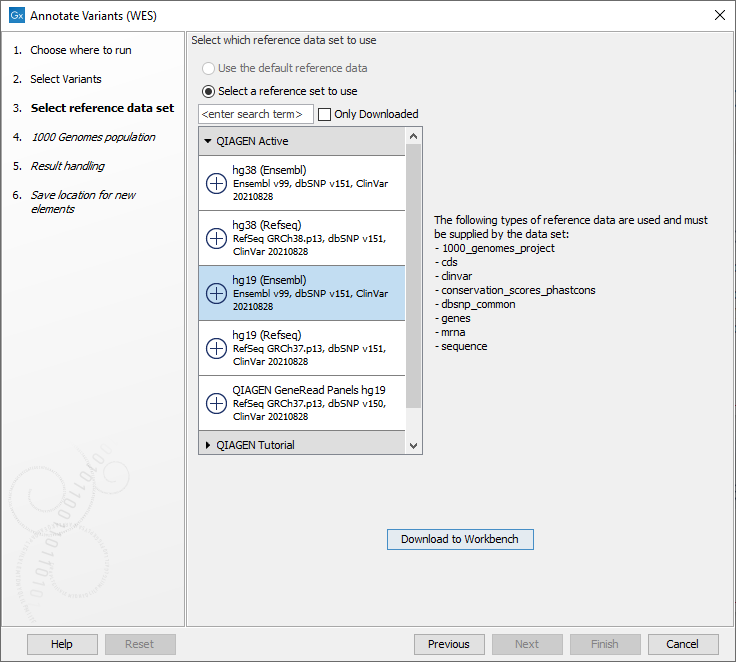
Figure 18.2: Choose the relevant reference Data Set to annotate. - The 1000 Genomes population(s) is bundled in the downloaded reference dataset and therefore preselected in this step. If you want to use another variant track you can browse in the navigation area for the preferred track to use (figure 18.3).
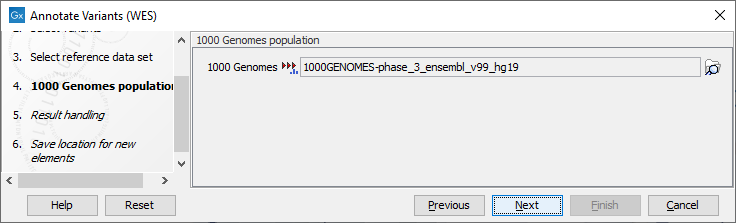
Figure 18.3: Use the preselected 1000 Genomes population(s) or select another variant track. - In the last wizard step you can check the selected settings by clicking on the button labeled Preview All Parameters.
In the Preview All Parameters wizard you can only check the settings, and if you wish to make changes you have to use the Previous button from the wizard to edit parameters in the relevant windows.
- Choose to Save your results and click on the button labeled Finish.
