Realignment of unaligned ends
A typical error in read alignments is the occurrence of unaligned ends (also known as soft-clipped read ends). These unaligned ends are introduced by the read mapper as a consequence of an unresolved indel towards the end of a read. Those unaligned ends can be realigned in many cases, after the read itself has been locally realigned according to the indel that prevented the read mapper from aligning the read ends correctly. Figure 27.34 depicts such an example.
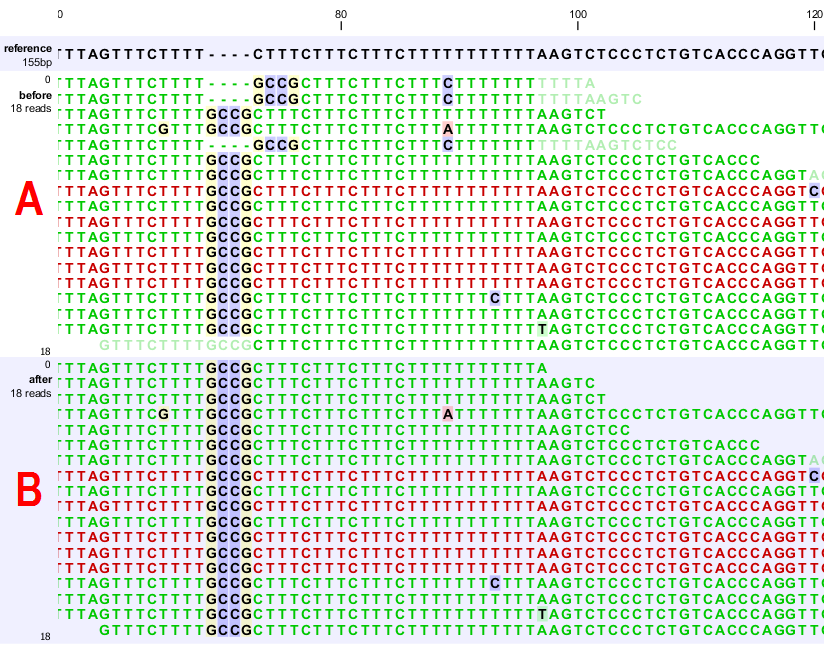
Figure 27.34: [A] The alignments of the first, second, and fifth read in this read mapping do not support the four-nucleotide insertion supported by the remaining reads. Additionally, the first, second, fifth and the last reads have unaligned ends. [B] After applying local realignment the first, second and fifth read consistently support the four-nucleotide insertion. Additionally, all previously unaligned ends have been realigned, because they perfectly match the reference sequence now (see also figure 27.31).
