Create RRBS-fragment Track
Create RRBS-fragment Track can be used to generate fragments of pre-selected size based on the restriction digest of a reference genome with commonly used frequent cutters targeting methylation contexts such as MspI. If such track is provided to the Call Methylation Levels tool, its features are used to calculate statistical tests instead of consecutive windows of pre-set size. The input object is a reference genome of interest, in a track format.
Figure 33.31 shows the options for generating a track with predicted, and selected features corresponding to restriction digest of the selected genome.
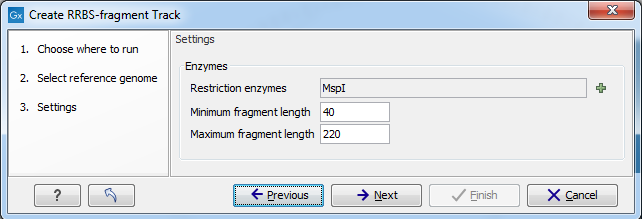
Figure 33.31: Create RRBS-fragment track settings.
Restriction enzymes The enzymes used in reduced-represention bisulfite sequencing can be selected here, with the most common for the CpG islands, MspI, pre-selected by default.
Parameters Minimum fragment length and Maximum fragment length define the acceptable size range, out of the all possible predicted fragments after full digest of reference DNA, will be included in the output of the tool.
The output of the tool is a feature track.
