Batch overview
The next Wizard step is the batch overview where you have the opportunity to refine the list of data that will be in each batch unit. For example, you could use this step to ensure that only trimmed sequence lists - and not all sequence lists - should be used for the analysis that is being setup.
The batch overview lists the batch units on the left and the contents of the selected batch unit on the right (figure 9.9).
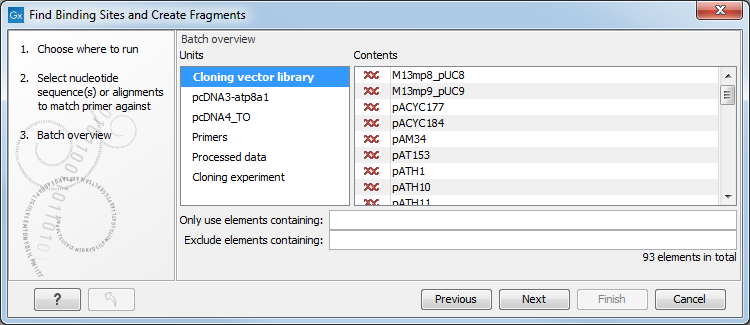
Figure 9.9: Overview of the batch run. At the bottom right, the number of files to be analysed, summed across all batch units, is shown, 92 in this case.
In this example, the two sequences (pcDNA) are defined as separate batch units because they are located at the top level of the Cloning folder. Of the four subfolders of the Cloning folder initially selected, three are listed in this view. In each of these subfolders, any data elements that the analysis could use as input will be used unless action is taken at this point to exclude some of these. So all the elements in the subfolder "Cloning vector library" and shown on the right-hand side of the dialog will be included as part of a single analysis run.
Note that folders that do not contain any data that can be used by the tool being launched will not be shown in that dialog.
Including and excluding data elements in batch units
There are three ways to refine the data elements that should be included in a batch unit, and thereby get taken forward into the analysis.
- Use the fields labeled Only use elements containing and Exclude elements containing at the bottom of the batch overview This refinement is done based on data element names. for example, only paired reads might be desired for the analysis, in which case, putting the text "paired" into the Only use elements containing field might be useful.
- Remove a whole batch unti Right-click on the batch unit to be removed and choose the option Remove Batch Unit.
- Remove a particular data element from a batch unit Right click on the element of a batch unit to be removed and choose the option Remove Element. This can be useful when filtering based on name, described in the first option, cannot be used to refine the batch units specifically enough.
