Create Variant Track Statistics Report
The tool Create Variant Track Statistics Report takes a variant track as input and reports statistics on variants classified by types. When the track contains the appropriate annotations, the report will also include sections on biological consequences and/or splice site effect.
To run the tool, go to:
Toolbox | Resequencing Analysis (![]() ) | Variant Quality Control (
) | Variant Quality Control (![]() ) | Create Variant Track Statistics Report (
) | Create Variant Track Statistics Report (![]() )
)
In the first dialog, select the variant track you want to get statistics for. In the second dialog you can optionally specify a second variant track, that is a filtered version of the input track. The tool will check that all variants present in the filtered track are also present in the input track. If it is the case, the report will give statistics "before filtering" (based on the input track) and "after filtering" (based on the optional filtered track). When the compatibility check fails, the filtered track will be ignored.
Variant types section
This section summarizes the count of non-reference variants of different types: SNV, MNV, Insertion, Deletion, and Replacement. Figure 29.15 shows the content of the section when only an input track is provided (left), and when both input and filtered tracks are provided (right).
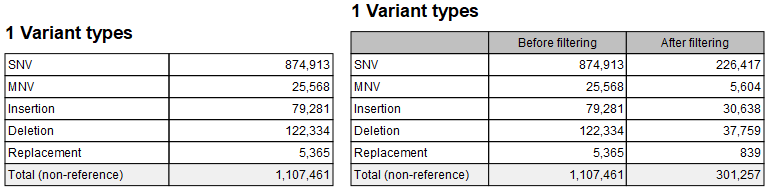
Figure 29.15: Variant types sections without (left) and with (right) a filtered track.
Amino acid changes section
This section summarizes the count of non-reference variants based on whether they are situated outside exons (so without any effect on amino acid change), and when exonic, whether the change is synonymous or not. To be present in the report, this section requires previous annotation of the variant track(s) with the Amino Acid Changes tool. Figure 29.16 shows the content of this section when a filtered track is provided. The example to the right shows what happens when the filtered track is missing annotations (statistics are then reported as Not Available).
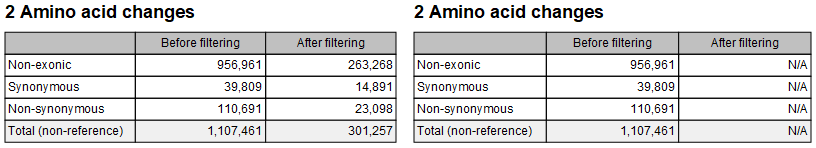
Figure 29.16: Amino acid changes sections with a filtered track that contains annotations (left), and with a filtered track missing relevant annotations (right).
Splice site effect section
This section summarizes the effect on splice sites produced by variants: Possible splice site disruption, and No splice site disruption. It requires previous annotation of the variant track(s) with the Predict Splice Site Effect tool. Figure 29.17 shows the content of this section when a filtered track was provided. The example to the right shows what happens when the input track misses the relevant annotations (statistics are then reported as Not Available). Note that the Predict Splice Site Effect tool only annotates variants that produce a possible splice site disruption. It is then possible that when no such variant is found, the annotated track is devoid of annotations, and the report section of the Create Variant Track Statistics Report tool will resemble the one obtained from a track that has not been annotated at all.
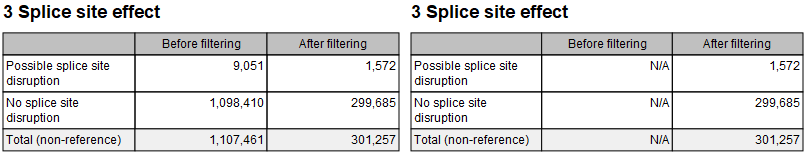
Figure 29.17: Splice site effect sections with a filter track that contains annotations (left), and with an input track missing relevant annotations (right).
