Output from Quantify QIAseq RNA
Two outputs are produced by the Quantify QIAseq RNA tool:
- Expression track. A track containing the genes included in the panel and their expression levels.
- Report. A report summarizing various QC metrics regarding which reads were ignored and which reads were included in a UMI read.
Expression track
The expression track produced by Quantify QIAseq RNA is displayed as a table listing genes included in the panel and several measures of their expression levels:
- Expression value. The number of distinct UMIs seen for this gene.
- TPM (Transcripts per Million). The number of transcripts per million that come from this gene. This is computed as the relative abundance per million
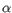 .
.
- RPKM (Reads Per Kilobase per Million reads). There is no good definition of RPKM for targeted amplicon data. We therefore define RPKM to be equal to TPM, which preserves the expected property that RPKM is proportional to TPM.
- Total gene reads. The number of distinct UMIs seen for this gene.
- Read count. The total number of reads mapping to this gene. Several reads may have the same UMI.
- UMI count. The number of distinct UMIs seen for this gene.
- Mean reads per UMI. The mean number of reads for each UMI seen for this target.
The expression track can be further analyzed using statistical tools from the RNA-Seq Analysis folder of the CLC Workbench, such as PCA for RNA-Seq and Create Feature Level Heat Map for RNA-Seq.
Report
The report produced by Quantify QIAseq RNA displays different QC metrics. They indicate how many reads were ignored and the reason they were not included in a UMI read. The "Accepted reads" column contains the number of reads that passed the filtering.
When you are designing a customized panel online, you will see that there is an option to add a set of 6 "GDC controls". These values are your negative controls of genomic DNA. You can also see these in the target regions where they have names like "GDC_CONTROL_06". The number of control targets with more than 10 UMI reads are listed in the "Expressed GDC controls" table cell. This cell will be colored pink and a warning will be shown if any controls are expressed.
Why does the tool not produce a read mapping?
The very short target regions in a QIAseq Targeted RNA Panel are not suited for downstream analyses that require a read mapping, such as variant calling. If a read mapping is desired, for example to investigate suspected off-target effects, we recommend using the Map Reads to Reference tool.
