Variant Detectors - overview
The Variant Detectors tools consist of three different tools for variant detection: The 'Basic Variant Detection' tool, the 'Fixed Ploidy Variant Detection' tool, and the 'Low Frequency Variant Detection' tool. The tools are designed for analysis of different types of samples. They differ in their underlying assumptions about the data, and hence differ in their assessments of when there is enough information in the data for a variant to be called. They share a number of options regarding which reads in the data should be considered, and options related to the filtering of called variants.
The Cancer and Genomics Workbenches offer three tools for detecting variants:
- The 'Basic Variant Detection' tool
- The 'Fixed Ploidy Variant Detection' tool and
- The 'Low Frequency Variant Detection' tool
The 'Basic Variant Detection' and the 'Fixed Ploidy Variant Detection' tools are new versions of the 'Quality-based Variant Detection' and the 'Probabilistic Variant Detection' tools, in which the filtering options have been unified and extended. The 'Quality-based Variant Detection' and the 'Probabilistic Variant Detection' are on their way to be retired and have been moved to the Legacy Tools.
The Basic Variant Detection, Fixed Ploidy Variant Detection and the Low Frequency Variant Detection tools differ in their underlying assumptions about the data, and are suitable for different types of applications. An overview is given in figure 27.10.
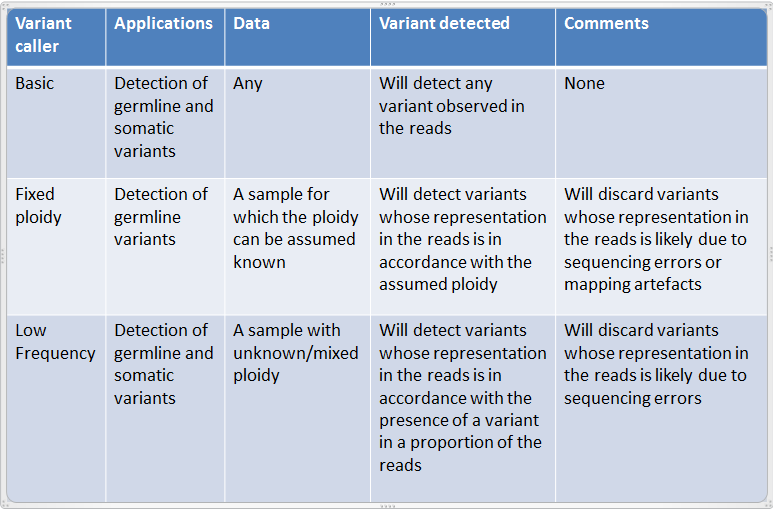
Figure 27.10: An overview of the applicabilities of the variant detection tools.
Below we first describe the tools. Each of the tools have a set of parameters that are specific to that tool. Next, we describe the filtering and output options that are shared among the tools (Filters).
To run the Variant Detection tools, go to:
Toolbox | Resequencing Analysis (![]() ) | Variant Detectors
) | Variant Detectors
Here you are presented with the three tools (see figure 27.11).
Figure 27.11: The Variant Detection tools.
When double-clicking one of the tools, a dialog is opened where you select the reads track or read mapping you want to analyze.
Figure 27.12: Select the read mapping that you want to analyze.
Click Next when the reads track is listed in the right-hand side of the dialog.
The user is next asked to set the parameters that are specific for the variant detection tool. The three tools, their assumptions, and the tool-specific parameters are described in the sections Basic Variant Detection, Fixed Ploidy Variant Detection, Low Frequency Variant Detection. First we will here give some examples of different variants called by the three variant detection tools and the overall mode by which the variant detection is carried out.
Subsections
