General filters
The 'General' filters relate to the regions and reads in the read mappings that should be considered, and the amount of evidence the user wants to require for a variant to be called:
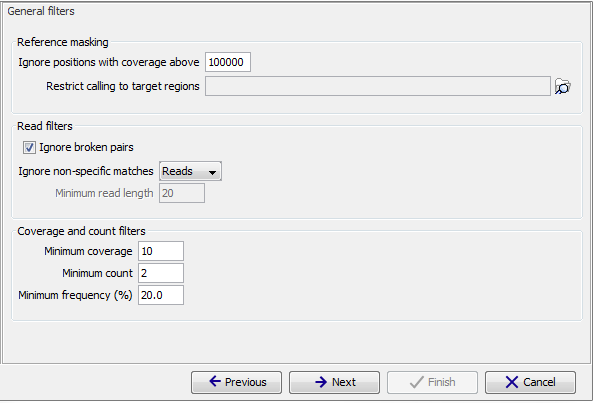
Figure 27.20: General filters. The values shown are those that are default for Fixed Ploidy Variant detection.
Note on the use of the rare variant caller with WGS data: The default settings for the rare variant caller are optimized for targeted resequencing protocols, not whole genome sequencing (e.g. cancer gene panels). In order to run the tool on WGS data the the parameter 'Ignore positions with coverage above' should be adjusted (to e.g. 1000) as it is not uncommon to have modest coverage for most part of the mapping, and abnormal areas (typically repeats around the centromeres) with very high coverage. Looking for low frequency variants in high coverage areas will exhaust the machine memory simply because there will be many low frequency variants (due to some reads originating from near identical repeat sequences or simple sequencing errors). As these 'high coverage' regions typically are not of interest, disregard these areas by using a lower 'Ignore positions with coverage above' filtering setting is recommended for WGS data.
Subsections
