Technology specific filters
- Remove pyro-error variants: This filter can be used to remove insertions and deletions in the reads that are likely to be due to pyro-like errors in homopolymer regions. There are two types of such errors: They may occur either at (1) the immediate ends of homopolymer regions or (2) as an 'overspill' a few nucleotides downstream of a homopolymer region. In case (1) the exact numbers of the same number of nucleotide is uncertain and a sequence like "AAAAAAAA" is sometimes reported as "AAAAAAAAA". In case (2) a sequence like "CGAAAAAGTCG" may sometimes get an 'overspill' insertion of an A between the T and C so that the reported sequence is C "CGAAAAAGTACG". Note that the removal is done in the reads as a very first step, before calling the initial 1 bp variants.
There are two parameters that must be specified for this filter:
- In homopolymer regions with minimum length: Only insertion or deletion variants in homopolymer regions of at least this length will be removed.
- With frequency below: Only insertion or deletion variants whose frequency (ignoring all non-reference and non-homopolymer variant reads) is lower than this threshold will be removed.
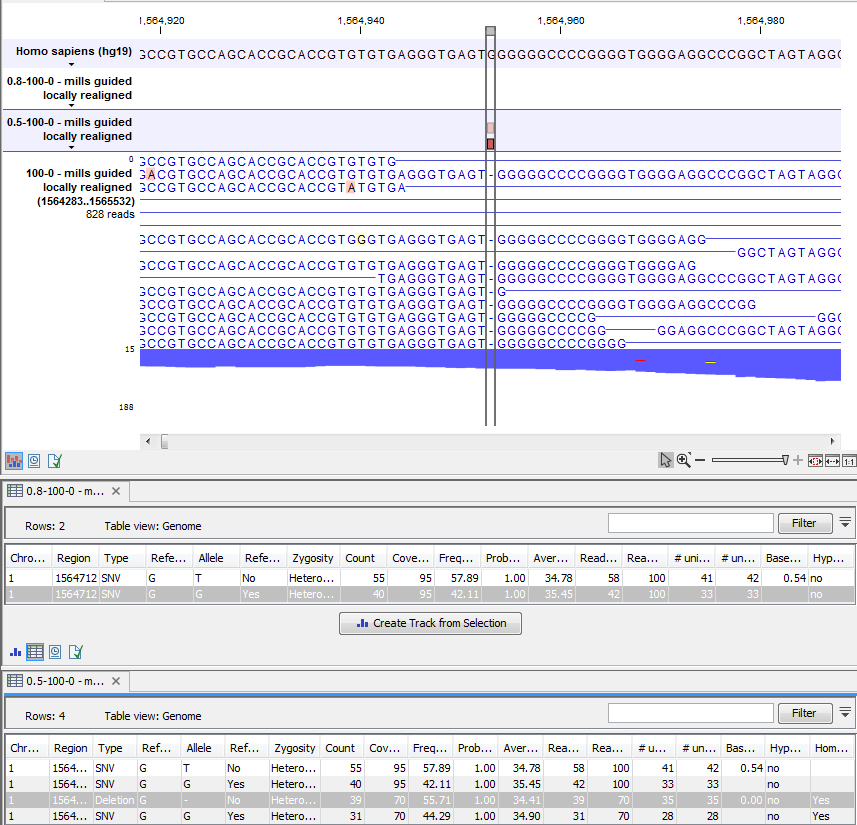
Figure 27.27: An example of a variant that is filtered out when the pyro-error filter is applied with settings 3 and 0.8, but not with settings 3 and 0.5.
In addition to the example above, a simple example is provided below in figure 27.28 to illustrate the difference between variant frequency and pyro-variant removal frequency (where non-reference and non-homopolymer variant reads are ignored).

Figure 27.28: An example of a simple read mapping with 6 mapped reads. Three of them indicate a deletion, two match the reference, and one read is an A to T SNP
The read with the T variant is not counted when calculating the frequency for the homopolymer deletion, because we only want to estimate how often a homopolymer variant appears for a given allele, and the T read is not from the same allele as the A and gap reads.
For the deletion, the variant frequency will be 50 percent, if it is reported. This is because it appears in 3 of 6 reads.
However, the pyro-variant removal frequency is 0.6, because it appears in 3 of 5 reads that come from the same allele. Thus the deletion will only be removed by the pyro-filter if the With frequency below parameter is above 0.6 (and the In homopolymer regions with minimum length parameter is less than 7).
