Direction and position filters
Many sequencing protocols are prone to various types of amplification induced biases and errors. The 'Read direction' and 'Read position' filters are aimed at providing means for weeding out variants that are likely to originate from such biases.
- Read direction filter: The read direction filter removes variants that are almost exclusively present in either forward or reverse reads. For many sequencing protocols such variants are most likely to be the result of amplification induced errors. Note, however, that the filter is NOT suitable for amplicon data, as for this you will not expect coverage of both forward and reverse reads. The filter has a single parameter:
- Direction frequency: Variants that are not supported by at least this frequency of reads from each direction are removed.
- Relative read direction filter: The relative read direction filter attempts to do the same thing as the 'Read direction filter', but does this in a statistical, rather than absolute, sense: it tests whether the distribution among forward and reverse reads of the variant carrying reads is different from that of the total set of reads covering the site. The statistical, rather than absolute, approach makes the filter less stringent. The filter has one parameter:
- Significance: Variants whose read direction distribution is significantly different from the expected with a test at this level, are removed. The lower you set the significance cut-off, the fewer variants will be filtered out.
- Read position filter: The read position filter is a filter that attempts to remove systematic errors in a similar fashion as the 'Read direction filter', but that is also suitable for hybridization-based data. It removes variants that are located differently in the reads carrying it, than would be expected given the general location of the reads covering the variant site. This is done by categorizing each sequenced nucleotide (or gap) according to the mapping direction of the read and also where in the read the nucleotide is found; each read is divided in five parts along its length and the part number of the nucleotide is recorded. This gives a total of ten categories for each sequenced nucleotide and a given site will have a distribution between these ten categories for the reads covering the site. If a variant is present in the site, you would expect the variant nucleotides to follow the same distribution. The read position filter carries out a test for whether the read position distribution of the variant carrying reads is different from that of the total set of reads covering the site. The filter has one parameter:
- Significance: Variants whose read position distribution is significantly different from the expected with a test at this level, are removed. The lower you set the significance cut-off, the fewer variants will be filtered out.
Figure 27.24 shows an example of a variant that is removed by the 'Read direction' filter.
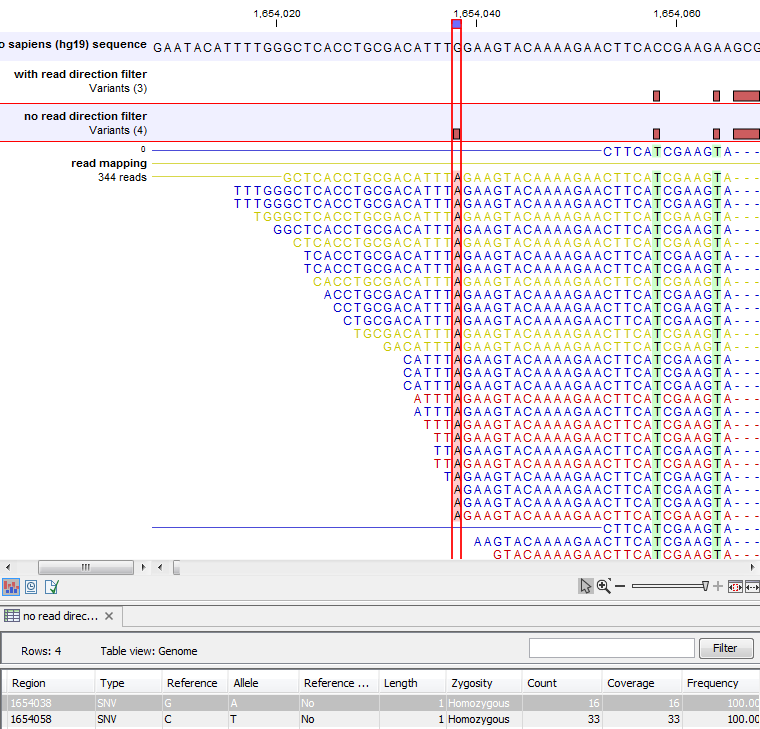
Figure 27.24: An example of a variant that is filtered out by the Read Direction filter.
Note that variant calling was done ignoring non-specific matches and broken pair reads, so only the 16 intact paired reads (the blue reads) are considered. To see the direction of the reads, you must adjust the viewer settings in the 'Reads track' side panel, to 'Disconnect paired reads'. This has been done in figure 27.25. Now it becomes apparent that the variant is found in the forward reads (that is, the green reads) of the 16 intact paired reads, and in no reverse reads (except the three that come from broken pairs, and which were ignored), and therefore removed by the read direction data.
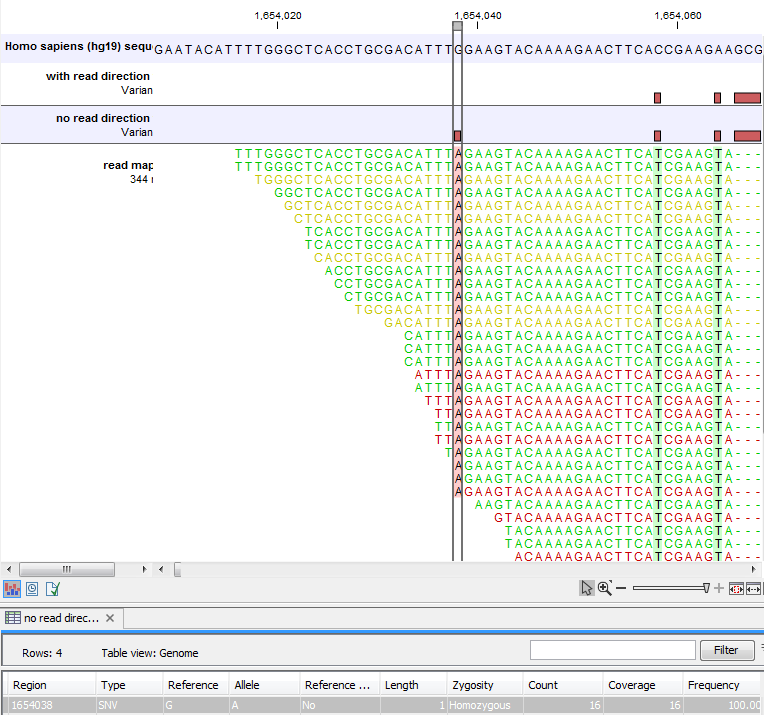
Figure 27.25: The same data as shown in figure 27.24, but now with 'Disconnect paired reads' option switched on in the 'reads track' side panel.
Figure 27.26 shows an example of a variant that is removed by the read position filter, but not by the read direction filter. The variant is only present in a portion of the reads that cover the variant, and the portion or the reads that carry the variant have the variant occurring in read positions that are systematically different from what you would expect, given the general placement of reads covering the variant (e.g., none of the reads that start after position 186,641,600 carry the variant).
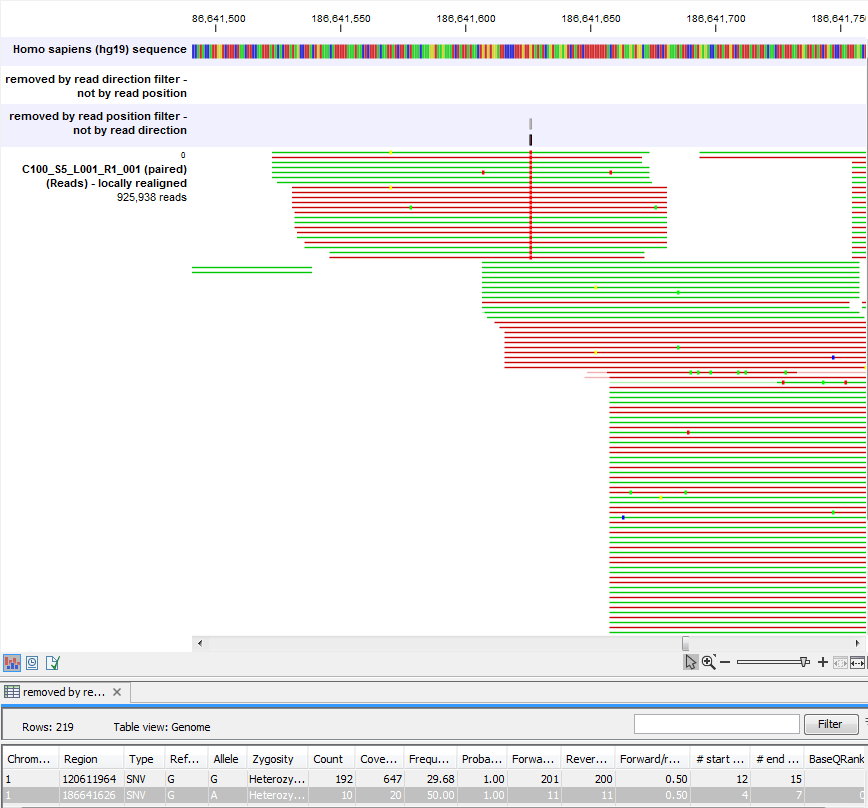
Figure 27.26: A variant that is filtered out by the Read position filter but not by the Read direction filter.
