View settings in the Side Panel
When you open a single mapping, the following settings are available in the Side Panel for customizing the layout.
- Read layout. A new preference group located at the top of the Side Panel:
- CompactnessThe compactness is an overall setting that lets you control the level of detail to be displayed on the sequencing reads. Please note that this setting affects many of the other settings in the Side Panel and the general behavior of the view as well. For example: if the compactness is set to Compact, you will not be able to see quality scores or annotations on the reads, no matter how this is specified in the respective settings. And when the compactness is Packed, it is not possible to edit the bases of any of the reads. There is a shortcut way of changing the compactness: Press and hold the Alt key while you scroll using your mouse wheel or touchpad.
- Not compact. The normal setting with full detail. If you wish to view trace data within the mapping, this option should be chosen. See also Trace settings in the Side Panel
- Low. Hides trace data, quality scores and puts the reads' annotations on the sequence.
- Medium. The labels of the reads and their annotations are hidden, and the residues of the reads cannot be seen.
- Compact. Even less space between the reads.
- Packed. All the other compactness settings will stack the reads on top of each other, but the packed setting will use all space available for displaying the reads. When zoomed in to 100%, you can see the residues but when zoomed out the reads will be represented as lines just as with the Compact setting. Please note that the packed mode is special because it does not allow any editing of the read sequences and selections, and furthermore the color coding that can be specified elsewhere in the Side Panel does not take effect. An example of the packed compactness setting is shown in figure 25.22.
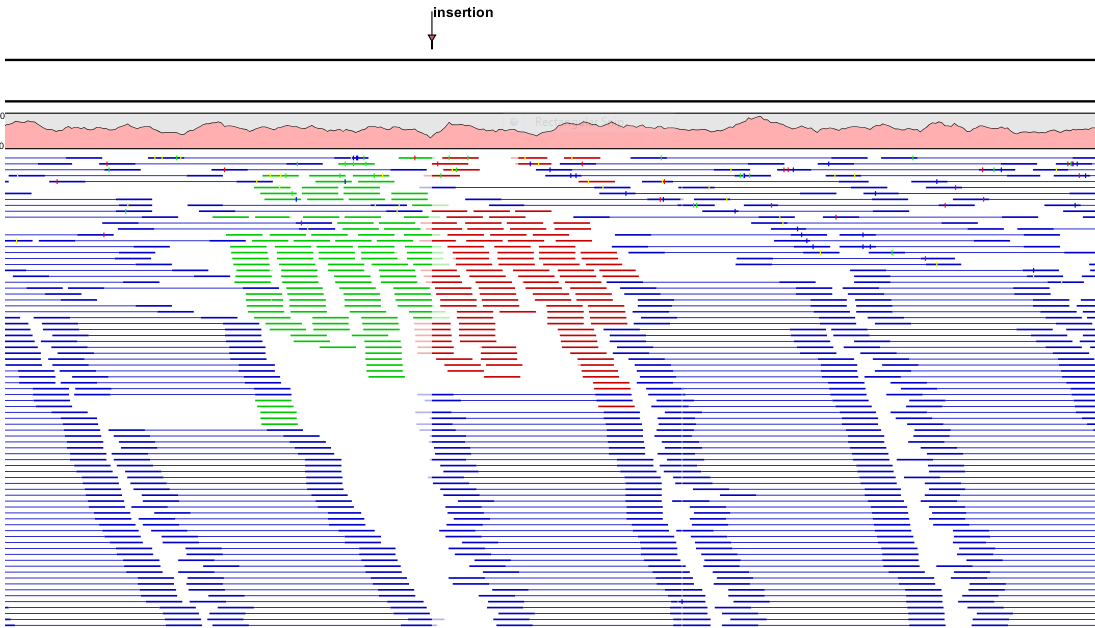
Figure 25.22: An example of the packed compactness setting. - Gather sequences at top. Enabling this option affects the view that is shown when scrolling horizontally. If selected, the sequence reads which did not contribute to the visible part of the mapping will be omitted whereas the contributing sequence reads will automatically be placed right below the reference. This setting is not relevant when the compactness is packed.
- Show sequence ends. Regions that have been trimmed are shown with faded traces and residues. This illustrates that these regions have been ignored during the assembly.
- Show mismatches. When the compactness is packed, you can highlight mismatches which will get a color according to the Rasmol color scheme. A mismatch is whenever the base is different from the reference sequence at this position. This setting also causes the reads that have mismatches to be floated at the top of the view.
- Disconnect pairs. This option will break up the paired reads in the display (they are still marked as pairs - this is just affects the visualization). The reads are marked with colors for the direction (default red and green) instead of the color for pairs (default blue). This is particularly useful when investigating overlapping pairs in packed view and when the strand / read orientation is important.
- Packed read height. When the compactness is set to "packed", you can choose the height of the visible reads. When there are more reads than the height specified, an overflow graph will be displayed below the reads. The overflow graph is shown in the same colors as the sequences, and mismatches in reads are shown as narrow horizontal lines in. The colors of the small lines represent the mismatching residue. The color codes for the horizontal lines correspond to the color used for highlighting mismatches in the sequences (red = A, blue = C, yellow = G, and green = T). E.g. a red line with half the height of the blue part of the overflow graph will represent a mismatching "A" in half of the paired reads at this particular position.
- Find Conflict. Clicking this button selects the next position where there is an conflict between the sequence reads. Residues that are different from the reference are colored (as default), providing an overview of the conflicts. Since the next conflict is automatically selected it is easy to make changes. You can also use the Space key to find the next conflict.
- Low coverage threshold. All regions with coverage up to and including this value are considered low coverage. When clicking the 'Find low coverage' button the next region in the read mapping with low coverage will be selected.
- CompactnessThe compactness is an overall setting that lets you control the level of detail to be displayed on the sequencing reads. Please note that this setting affects many of the other settings in the Side Panel and the general behavior of the view as well. For example: if the compactness is set to Compact, you will not be able to see quality scores or annotations on the reads, no matter how this is specified in the respective settings. And when the compactness is Packed, it is not possible to edit the bases of any of the reads. There is a shortcut way of changing the compactness: Press and hold the Alt key while you scroll using your mouse wheel or touchpad.
- Alignment info. There is one additional parameter:
- Coverage: Shows how many sequence reads that are contributing information to a given position in the mapping. The level of coverage is relative to the overall number of sequence reads.
- Foreground color. Colors the letters using a gradient, where the left side color is used for low coverage and the right side is used for maximum coverage.
- Background color. Colors the background of the letters using a gradient, where the left side color is used for low coverage and the right side is used for maximum coverage
- Graph. The coverage is displayed as a graph (Learn how to export the data behind the graph).
- Height. Specifies the height of the graph.
- Type. The graph can be displayed as Line plot, Bar plot or as a Color bar.
- Color box. For Line and Bar plots, the color of the plot can be set by clicking the color box. If a Color bar is chosen, the color box is replaced by a gradient color box as described under Foreground color.
- Coverage: Shows how many sequence reads that are contributing information to a given position in the mapping. The level of coverage is relative to the overall number of sequence reads.
- Residue coloring. There is one additional parameter:
- Sequence colors. This option lets you use different colors for the reads.
- Main. The color of the consensus and reference sequence. Black per default.
- Forward. The color of forward reads (single reads). Green per default.
- Reverse. The color of reverse reads (single reads). Red per default.
- Paired. The color of paired reads. Blue per default. Note that reads from broken pairs are colored according to their Forward/Reverse orientation or as a Non-specific match, but with a darker nuance than ordinary single reads.
- Non-specific matches. When a read would have matched equally well another place in the mapping, it is considered a non-specific match. This color will "overrule" the other colors. Note that if you are mapping with several reference sequences, a read is considered a double match when it matches more than once across all the contigs/references. A non-specific match is yellow per default.
- Sequence colors. This option lets you use different colors for the reads.
Beside from these preferences, all the functionalities of the alignment view are available. This means that you can e.g. add annotations (such as SNP annotations) to regions of interest.
However, some of the parameters from alignment views are set at a different default value in the view of contigs. Trace data of the sequencing reads are shown if present (can be enabled and disabled under the Nucleotide info preference group), and the Color different residues option is also enabled in order to provide a better overview of conflicts (can be changed in the Alignment info preference group).
- Sequence layout. At the top of the Side Panel:
- Matching residues as dots Matching residues will be presented as dots. Only the top sequence will be preserved in its original format.
