Exon identification and discovery
Clicking Next will show the dialog in figure 27.7.
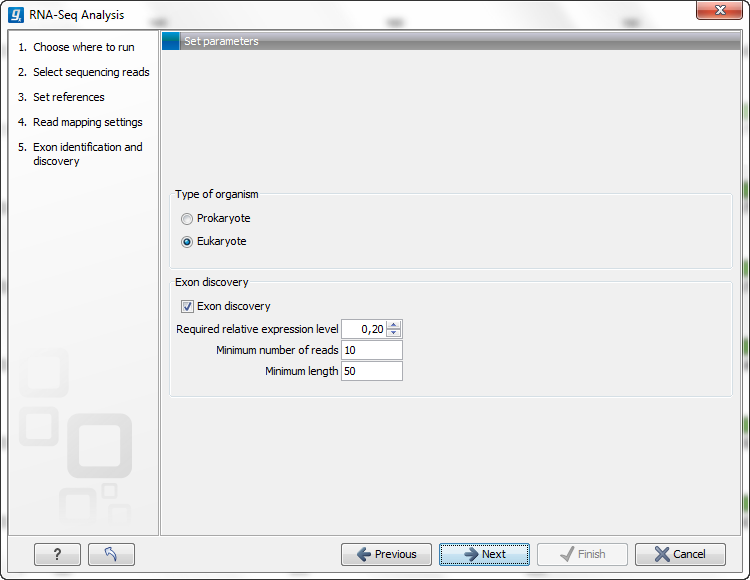
Figure 27.7: Exon identification and discovery.
The choice between Prokaryote and Eukaryote is basically a matter of telling the Workbench whether you have introns in your reference. In order to select Eukaryote, you need to have reference sequences with annotations of the type mRNA (this is the way the Workbench expects exons to be defined - see in the introduction).
Here you can specify the settings for discovering novel exons. The mapping will be performed against the entire gene, and by analyzing the reads located between known exons, the CLC Genomics Workbench is able to report new exons. A new exon has to fulfill the parameters you set:
- Required relative expression level. This is the expression level relative to the rest of the gene. A value of 20% means that the expression level of the new exon has to be at least 20% of that of the known exons of this gene.
- Minimum number of reads. While the previous option asks for the percentage relative to the general expression level of the gene, this option requires an absolute value. Just a few matching reads will already be considered to be a new exon for genes with low expression levels. This is avoided by setting a minimum number of reads here.
- Minimum length. This is the minimum length of an exon. There has to be overlapping reads for the whole minimum length.
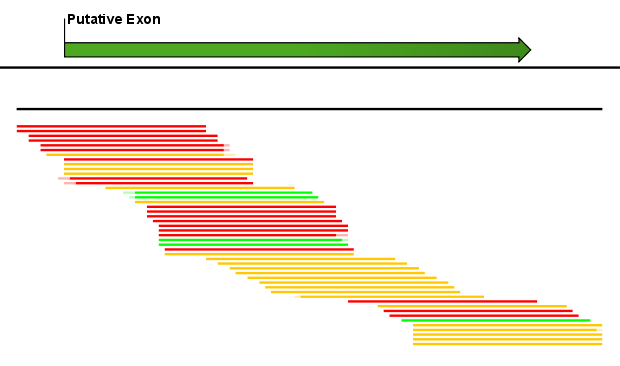
Figure 27.8: A putative exon has been identified.
