Pfam search parameters
- Choose database and search type
When searching for Pfam domains it is possible to choose different databases and specify the search for full domains or fragments of domains. Only the 100 most frequent domains are included as default in CLC Genomics Workbench. Additional databases can be downloaded directly from CLC bio's web-site at http://www.clcbio.com/resources.- Search full domains and fragments. This option allows you to search both for full domain but also for partial domains. This could be the case if a domain extends beyond the ends of a sequence
- Search full domains only. Selecting this option only allows searches for full domains.
- Search fragments only. Only partial domains will be found.
- Database. Only the 100 most frequent domains are included as default in CLC Genomics Workbench, but additional databases can be downloaded and installed as described in section Download and installation of additional Pfam databases.
- Set significance cutoff. The E-value (expectation value) is the number of hits that would be expected to have a score equal to or better than this value, by chance alone. This means that a good E-value which gives a confident prediction is much less than 1. E-values around 1 is what is expected by chance. Thus, the lower the E-value, the more specific the search for domains will be. Only positive numbers are allowed.
Click Next if you wish to adjust how to handle the results. If not, click Finish. This will open a viewshowing the found domains as annotations on the original sequence (see figure 16.17). If you have selected several sequences, a corresponding number of views will be opened.
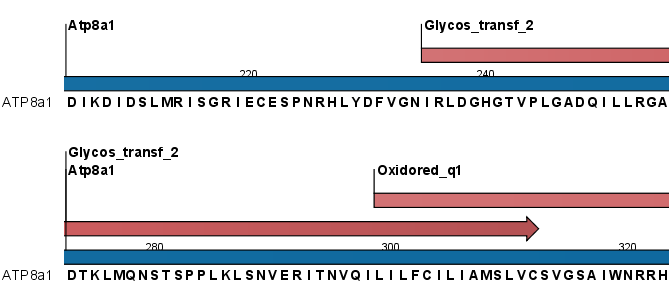
Figure 16.17: Domains annotations based on Pfam.
Each found domain will be represented as an annotation of the type Region. More information on each found domain is available through the tooltip, including detailed information on the identity score which is the basis for the prediction.
For a more detailed description of the provided scores through the tool tip look at http://pfam.sanger.ac.uk/help#tabview=tab5.
