Mapping parameters
Clicking Next leads to the parameters for the read mapping (see figure 25.2).
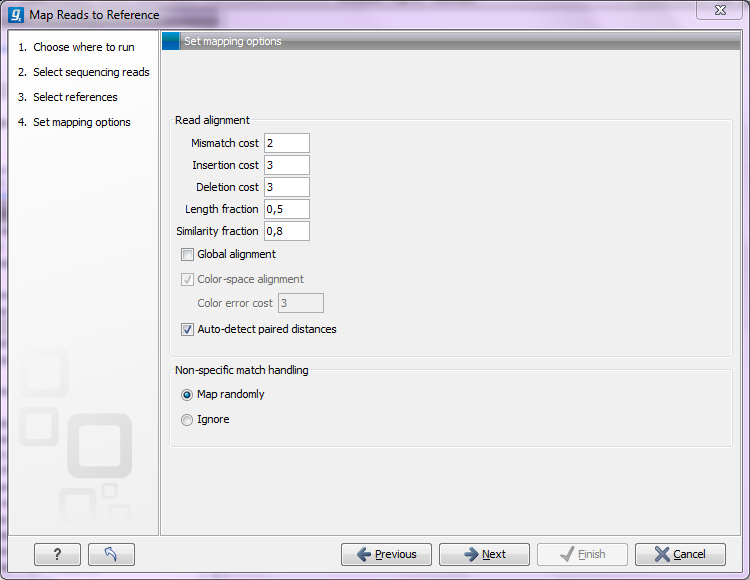
Figure 25.2: Setting parameters for the mapping.
At the top, you specify mismatch and gap costs:
- Mismatch cost
- The cost of a mismatch between the read and the reference sequence.
- Insertion cost
- The cost of an insertion in the read (causing a gap in the reference sequence)
- Deletion cost
- The cost of having a gap in the read.
Once the optimal alignment of the read is found, based on the costs specified above (e.g. to favor mismatches over indels), a filtering process determines whether this match is good enough for the read to be included in the output. The filtering threshold is determined by two fractions:
- Length fraction
- Set minimum length fraction of a read that must match the reference sequence. Setting a value at 0.5 means that at least half the read needs to match the reference sequence for the read to be included in the final mapping.
- Similarity
- Set minimum fraction of identity between the read and the reference sequence. If you want the reads to have e.g. at least 90% identity with the reference sequence in order to be included in the final mapping, set this value to 0.9. Note that the similarity fraction does not apply to the whole read; it relates to the Length fraction. With the default values, it means that at least 50 % of the read must have at least 90 % identity.
By default, mapping is done with local alignment of the reads to the reference. The advantage of performing local alignment instead of global alignment is that the ends are automatically left unaligned if there are many differences from the reference at the ends. For many sequencing platforms, the quality of the bases drop along the read, and a local alignment approach is desirable. Note that the aligned region has to be greater than the length threshold set. If global alignment is preferred, it can be enabled with a checkbox as shown in in figure 25.2.
When mapping data in color space (data from SOLiD systems), the color space checkbox is enabled, and a corresponding cost for color errors can be set. If you do not have color space data, these will be disabled and are not relevant. For more details about this, please see the section on Color space which explains how color space mapping is performed in greater detail.
Subsections
