Creating Left- and Right breakpoint signatures
There are typically numerous reads with unaligned ends in read mappings -- some are due to structural variants in the sample relative to the reference, others are due to poorly mapped, or poor quality reads. An example is given in figure 26.21. In order to make reliable predictions, attempts must be made to distinguish the unaligned ends caused by noisy read(mappings) from those caused by structural variants, so that the signal from the structural variants comes through as clearly as possible -- both in terms of where the 'significant' unaligned ends are and in terms of what they look like.

Figure 26.21: Example of a read mapping containing unaligned ends with three unaligned end signatures.
To identify positions with a 'significant' portion of 'consistent' unaligned end reads we first estimate 'null-distributions' of the fractions of left and right unaligned end reads at each position in the read mapping, and subsequently use these distributions to identify positions with an 'excess' of unaligned end reads. In these positions we create a Left (LB) or Right (RB) breakpoint signature. To estimate the null-distributions we:
- Calculate the coverage,
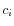 , in each position,
, in each position, 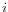 of all uniquely mapped reads (for paired read data sets, only intact paired reads pairs are considered -- broken paired reads are ignored).
of all uniquely mapped reads (for paired read data sets, only intact paired reads pairs are considered -- broken paired reads are ignored).
- Calculate the coverage in each position of 'valid' reads with a starting left unaligned end,
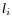 (of minimum consensus length 3bp).
(of minimum consensus length 3bp).
- Calculate the coverage in each position of 'valid' reads with a starting right unaligned end,
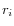 (of minimum consensus length 3bp).
(of minimum consensus length 3bp).
There are two user-specified settings, which control the significance of the LBs and RBs: 'The P-value threshold' and the 'Maximum number of mismatches' (see figure 26.19). The p-value is used as a cutoff in the binomial distributions estimated above: if the probability of obtaining the observed number of left (or right) unaligned ends in a position with the observed coverage, is smaller than the user-specified cut-off, a Left breakpoint signature (LB), respectively Right breakpoint signature (RB), is created. The 'Maximum number of mis-matches' parameter is used to determine which reads are considered 'valid' unaligned end reads. Only reads that have at most this number of mis-matches in their aligned parts are counted. The higher these two values are set, the more breakpoints will be called. The more breakpoints are called, the larger the search space for the Structural variation detection algorithm, and thus the longer the computation time.
In figure 26.21, three unaligned end signatures are shown. The left-most LB signature is called only when the p-value cut-off is chosen high (0.01 as opposed to 0.0001).
The 'Filter variants' parameter allows the user to filter out structural variants that derived from breakpoints that are supported by few reads. The number of reads supporting a structural variant is defined as the sum of the number of reads that support the breakpoints used to define the structural variant. Structural variants whose breakpoints are supported by fewer than the user specified cut-off are ignored.
