Predicting Structural Variants
Having created breakpoint signatures (LBs and RBs), we use these in a procedure, which inspects the called breakpoint signatures, and attempts to match and combine them to infer possible underlying structural variant signatures. Based on the inferred structural variant signatures, structural variants are predicted, and annotations created. The procedure for inspecting breakpoint signatures and inferring structural variants is described in detail below.
The 'Indels and Structural variants tool' has a number of outputs. The user may choose to have a report created. The report summarizes the number of breakpoints detected, provides some characteristics of the breakpoint, and on the numbers and types of structural variants detected. In addition to the report, the user may specify to have (1) the breakpoints, (2) the InDels and (3) the structural variants reported. These can be reported either as tracks or as tables, depending on the users choice. When the track option is chosen, the breakpoints and structural variants are reported in feature tracks, and the InDels in a variant track. The InDel track contains the small to medium sized insertions and deletions (shorter than approximately 220 bp) and for which the algorithm was able to identify the allele sequence (that is, the exact inserted sequence, or the exact deleted sequence).
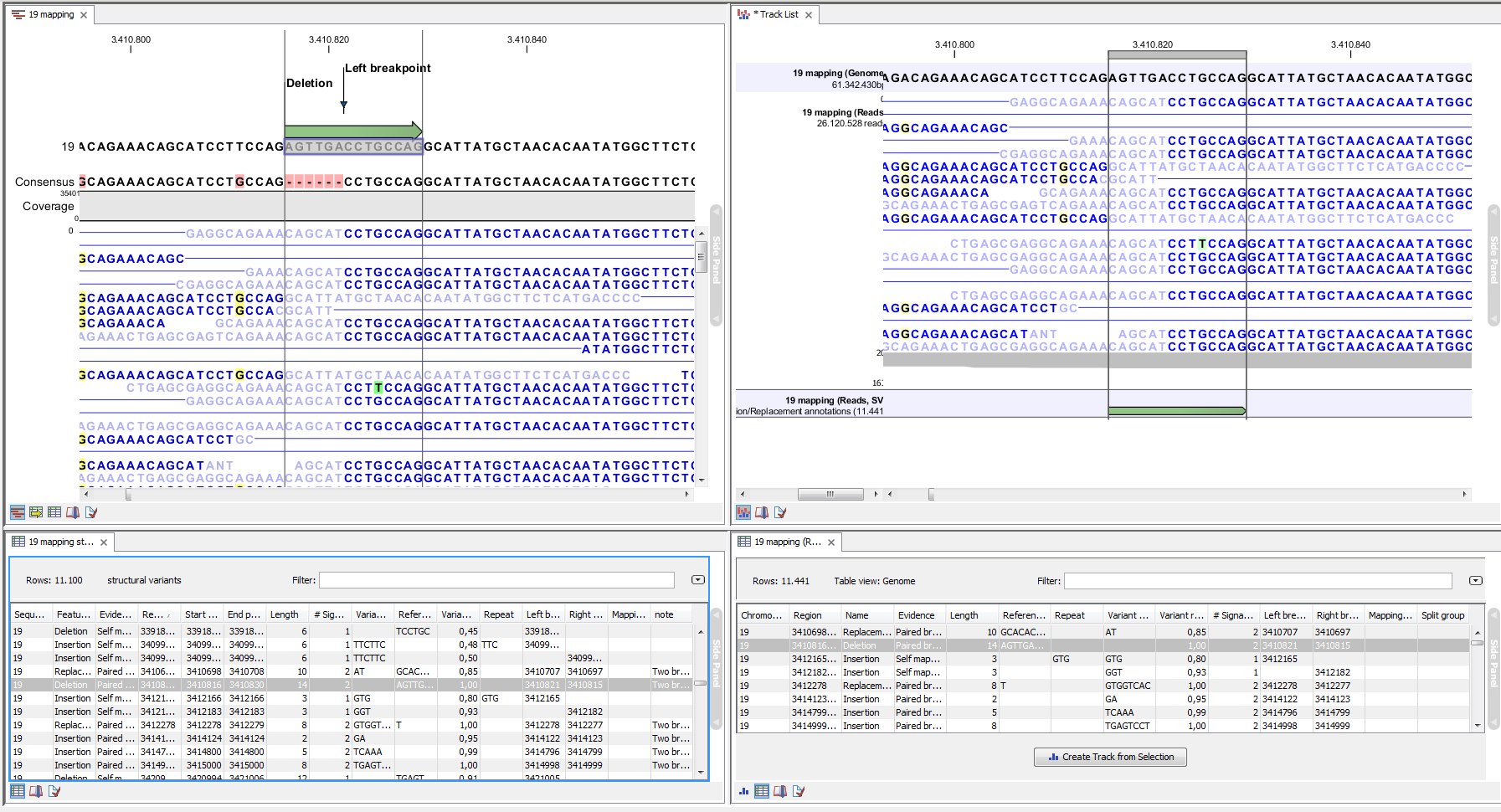
Figure 26.22: Example of the result of an analysis on a standalone read mapping (to the left) and on a reads track (to the right).
Typically, there will be called breakpoint signatures that are not found to stem from a structural variant. There may be a number of reasons for that: (1) the unaligned ends from which the breakpoint signature was derived might not be caused by an underlying structural variant, but merely be due to read mapping issues or noise, or (2) the breakpoint(s) which the detected breakpoint should have been matched to was/were not detected, and therefore no matching breakpoint(s) were found. Breakpoints may go un-detected either because of lack of coverage in the breakpoint region or because they are located within regions with exclusively non-uniquely mapped reads (only unaligned ends of uniquely mapping reads are used).
Subsections
