BLAST search
CLC Genomics Workbench offers to conduct BLAST searches on protein and DNA sequences. In short, a BLAST search identifies homologous sequences between your input (query) query sequence and a database of sequences[McGinnis and Madden, 2004]. BLAST (Basic Local Alignment Search Tool), identifies homologous sequences using a heuristic method which finds short matches between two sequences. After initial match BLAST attempts to start local alignments from these initial matches.
If you are interested in the bioinformatics behind BLAST, there is an easy-to-read explanation of this in Bioinformatics explained: BLAST.
With CLC Genomics Workbench there are two ways of performing BLAST searches: You can either have the BLAST process run on NCBI's BLAST servers (http://www.ncbi.nlm.nih.gov/) or perform the BLAST search on your own computer. The advantage of running the BLAST search on NCBI servers is that you have readily access to the most popular BLAST databases without having to download them to your own computer. The advantage of running BLAST on your own computer is that you can use your own sequence data, and that this can sometimes be faster and more reliable for big batch BLAST jobs
Figure 12.8 shows an example of a BLAST result in the CLC Genomics Workbench.
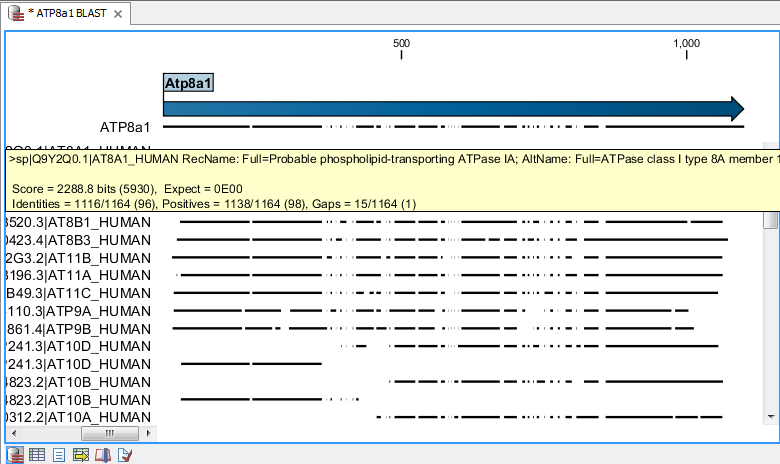
Figure 12.1: Display of the output of a BLAST search. At the top is there a graphical representation of BLAST hits with tool-tips showing additional information on individual hits. Below is a tabular form of the BLAST results.
Subsections
- Running BLAST searches
- Output from BLAST searches
- Local BLAST databases
- Manage BLAST databases
- Bioinformatics explained: BLAST
