Selecting data types for download
Once you have clicked Next, you will be asked whether you wish to download the genome sequence or whether you already have it available as shown in figure 11.7:
Figure 11.7: Selecting a reference genome sequence if available.
If you do not already have an existing sequence imported into the CLC Genomics Workbench, it will be downloaded automatically from Ensembl. If you already have a reference sequence, it has to match the genome definition built into the download tool. This means that the name and length of the chromosomes in your reference sequence have to match the genome definition of the tool.
Clicking Next allows you to select which types of annotation data you wish to download as shown in figure 11.8:
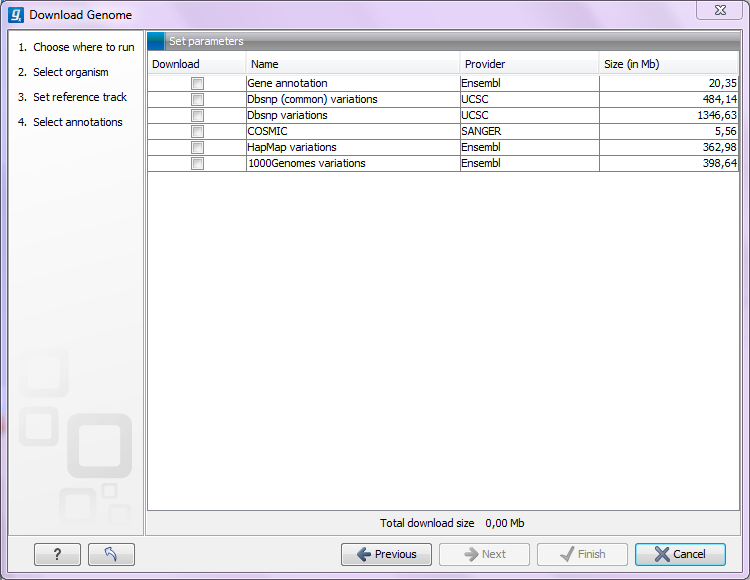
Figure 11.8: Selecting different types of annotation data for human hg19. .
This step is different from organism to organism and depends on the data sources that CLC bio has included for download. In the example in figure 11.8 showing hg19 build of the human genome, a lot of variant data is available from e.g. dbSNP or COSMIC. Please note that for human data, a difference between the UCSC genome build and Ensembl/NCBI exists, which means that variants downloaded from UCSC will not be annotated on the mitochondrial genome when using this download tool (see http://genome.ucsc.edu/cgi-bin/hgGateway?db=hg19).
Both the data provider and the size of the data is listed in the dialog, and at the bottom you can see the size of all the selected downloads. Please note that the file size displayed in the setup window for the Download Genomes tool refers to the size of the compressed text files, which the tool is retrieving from the provider's depository. The size of the track objects will be, after decompression and conversion from text to the .clc track format, notably larger.
All data downloaded with this tool will be tracks (either sequence tracks or various kinds of annotation tracks).
