Visualize drug interaction
Clicking a link provided in the 'Drug interaction in protein 3D structure' column will open a list with information about the drug hits (figure 30.35).
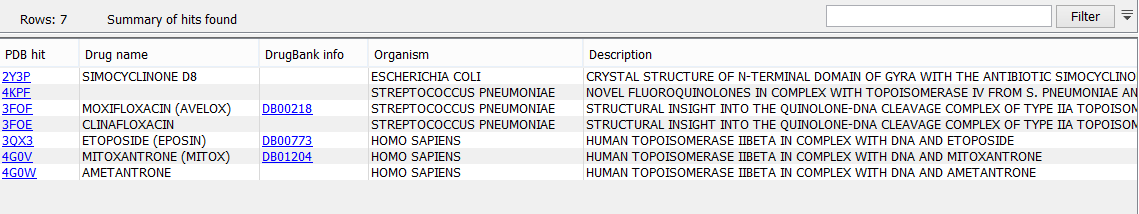
Figure 30.35: An example of a drug hit table with information about the drug and with links to 3D visualizations of variant-drug interaction.
The information available in the drug hit list is the following:
- PDB hit. Clicking a link provided in the PDB hit column will show a menu with two options: "Download and Show Structure" and "Help". "Help" gives access to this documentation. The "Download and Show Structure" option does exactly as described in Create 3D visualization of variant, except that the final 3D visualization is centered on the drug, and the drug is shown in ball n' sticks representation with atoms colored according to their atom types (figure 30.36).
- PDB drug name. (Hidden by default) The identifier used by PDB for the ligand or drug.
- Drug name. When possible, a common name for the potential drug is listed here. The name is taken from the corresponding DrugBank entry (if available) or from the PDB header information for the PDB hit.
- DrugBank info. If information about the drug or ligand is available in DrugBank (www.drugbank.ca [Law et al., 2014,Wishart et al., 2006]), a weblink to the appropriate site is listed here.
- E-value. (Hidden by default) The E-value is a measure of the quality of the match returned from the BLAST search. The closer to zero, the more homologous is the template structure to the query sequence.
- Organism. (Hidden by default) The organism for which the PDB structure has been obtained.
- Description. The description of the PDB file content, as given in the header of the PBD hit.
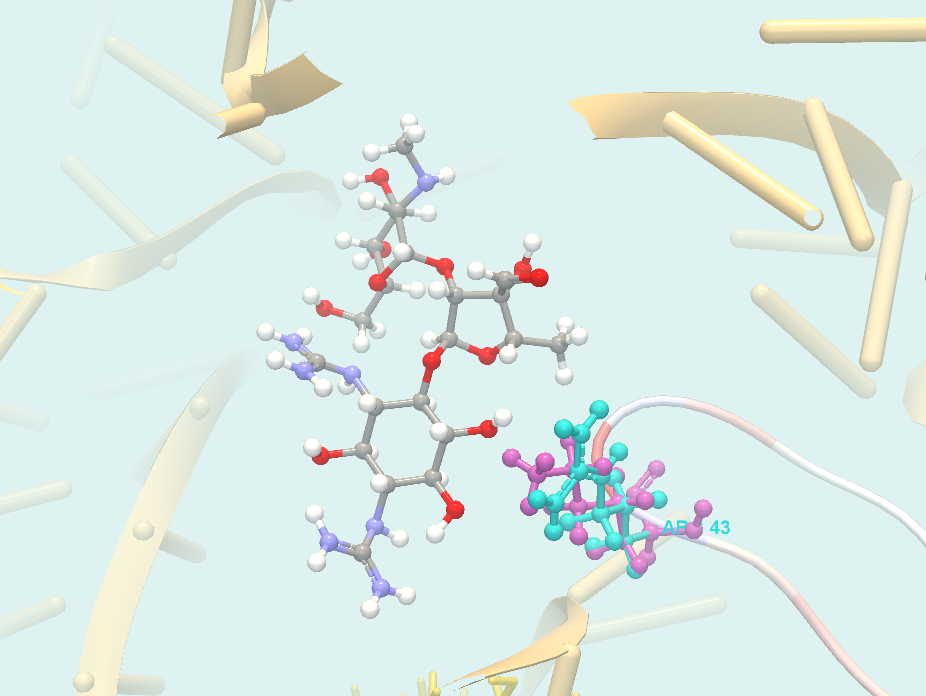
Figure 30.36: 3D visualization of variant-drug interaction. The drug (streptomycin) is in center of the view, and colored according to it's atom types. In this picture, the foreground has been cut away using the clipping plane functionality found in the Visualization tab in the Side Panel, to better see the variant-drug interaction.
To learn more about model structure, see Model structure.
