Download Genomes
Under the Download Genomes tab of the Reference Data Manager, you can access genomes and associated genomic data such as annotations and known variants from public repositories (figure 9.4). The data is not provided nor hosted by QIAGEN.
The list of organisms is kept up to date automatically. Click on an organism to select it. Information about the data available for download is then shown in the right hand panel. This includes a name describing the data, the data provider, a version where available, and the size.
If data for that organism has been downloaded previously, information about that is shown at the bottom of this area, under "Previous downloads".
Most data is supplied as a compressed text file. After download, each file is decompressed and the information imported into a track element (CLC format). CLC data is compressed by default, but the file size after import will generally not be the same as that reported in the Download Genomes tool.
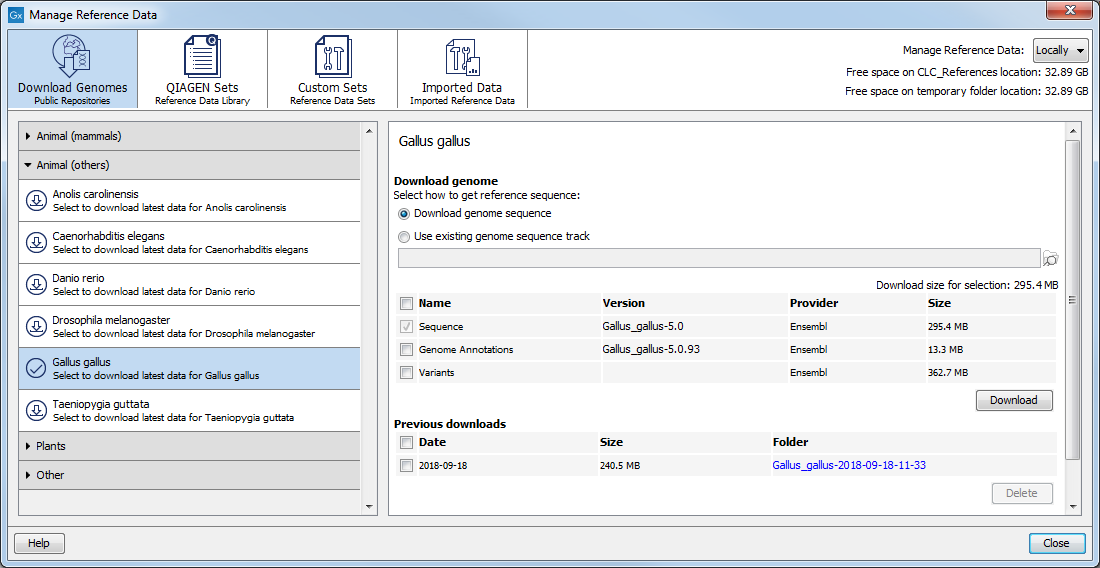
Figure 9.4: Download genomes and associated data for selected organisms.
The tracks imported are saved under the CLC_References location. A folder for each set downloaded is created under the "Genomes" folder. Its name contains the species name and the date of the download.
To delete data elements that have been downloaded, check the box beside the set of data to be deleted in the "Previous downloads" section of the right hand panel of the Reference Data Manager. The full set of data downloaded on a particular date will then be deleted.
When reference data is stored on a CLC Server, you need be logged in from the Workbench as an administrative user to be able to delete reference data.
Notes about particular data types
When GFF3 files are imported, a track is created for each feature type present in the file (see here).
In addition, an (RNA) track and a (Gene) track are created. The (RNA) track contains entries for all "RNA" type annotations. I.e. all the children of "mature_transcript", which is the parent of "mRNA", which is the parent of the "NSD_transcript". The (Gene) track contains genes and gene-like annotation types, such as ncRNA_gene, plastid_gene, and tRNA_gene. These broader sets of annotations can make these tracks particularly useful for some types of analyses, e.g. RNA-Seq.
For some genomes, chromosome bands (ideograms) are available (figure 9.5).
Please note that hg18 and hg19 variants downloaded from UCSC do not include variants on the mitochondrial genome.
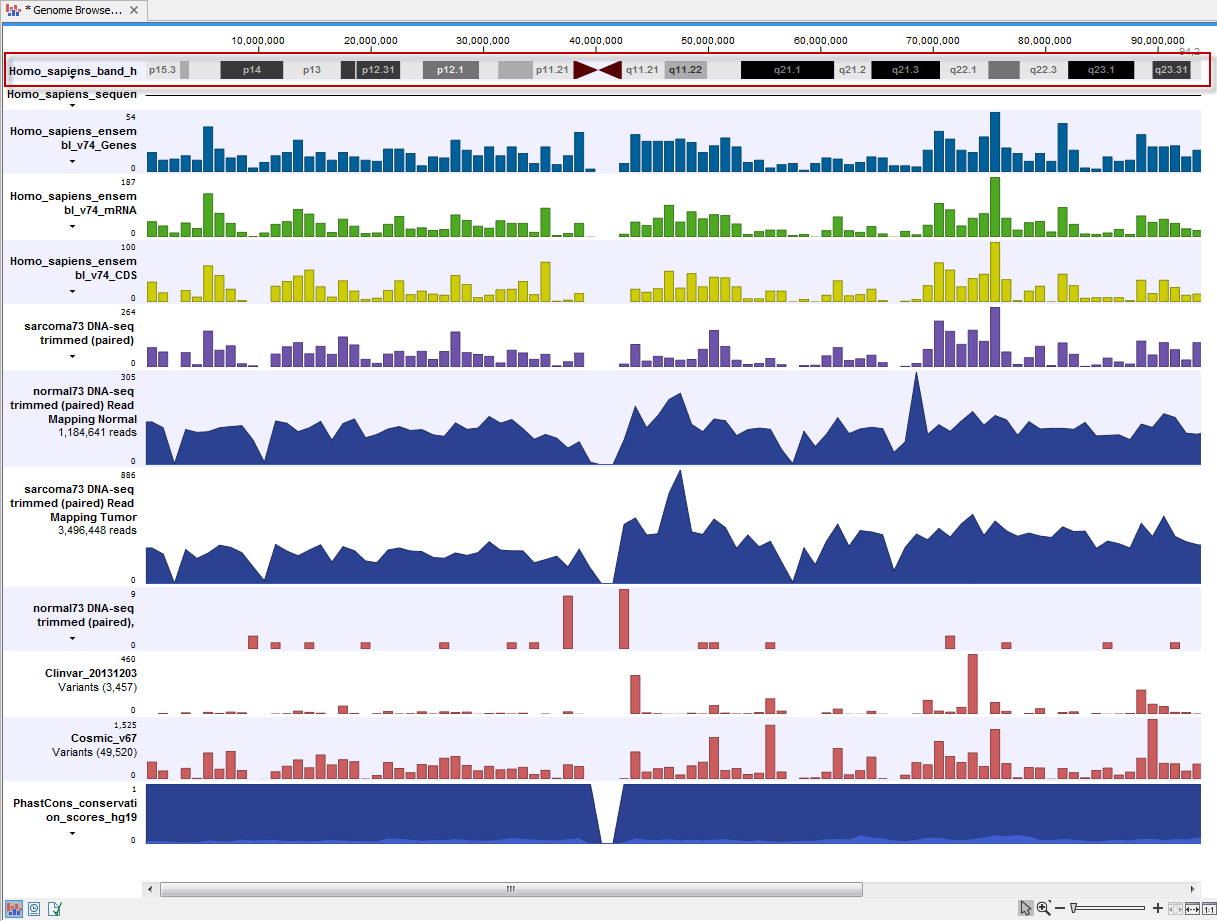
Figure 9.5: The ideogram is particularly useful when used in combination with other tracks in a track list. In this figure the ideogram is highlighted with a red box.
