Homopolymer trimming
Configuration for the homopolymer trimming step is shown in figure 26.12.
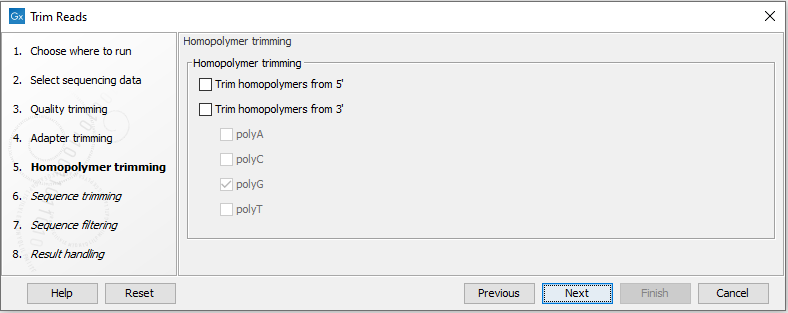
Figure 26.12: Homopolymer trimming.
Homopolymer trimming takes place only if at least one read end type is selected. After selecting the read end(s) to trim, you can select the type of homopolymer stretches to be removed.
How it works
Trimming of each type of homopolymer at each read end is done in the same way.
Using polyG as an example:
- A window of 10 nucleotides at the end of the read is initially checked.
- If fewer than 9 bases are Gs, then checking stops and no bases are trimmed.
- If all 10 bases are Gs, they are marked for trimming.
- If 9 out of 10 bases are Gs, all 10 bases are marked for trimming unless the non-G base is at the end of the 10 bases. In the following examples, where trimming takes place from left to right, the only base that is not marked for trimming is in bold:
- NGG GGG GGG G
- GGG GGN GGG G
- GGG GGG GGN G
- GGG GGG GGG N
The window then slides by one position, to cover 9 of the original bases and 1 additional base, and the steps decribed above are repeated.
This process continues until the sliding 10-base window contains fewer than 9 Gs. At that point, checking stops and all bases marked to be trimmed are removed.
Examples of the effects of trimming particular sequences:
- Trimming the sequence
CCCCCCTCCCCCCATATATATATATATCCCCCCTCCCCCfor polyC at the start and end of the read would result inATATATATATATAT. - Trimming the sequence
CCCCCCTCCCCCCATATATATATATATTTTTTTTTTGTTTTTTfor polyT and polyC at the start and end of the read would result inATATATATATA. Note that aTAis removed at the 3' end. This is because the 10-base windowTATTTTTTTTcontains nine Ts, and thus all 10 bases in this stretch are removed. - Trimming the sequence
AAAAAAAAAATATTTTTTTTTTGTTTTTTfor polyA and polyT at the start and end of the read would result in the whole sequence being trimmed away.
