Extract Annotated Regions
Using Extract Annotated Regions, parts of a sequence (or several sequences) can be extracted based on annotations. Lengths of flanking regions can be specified if desired. The output is a sequence list that contains sequences carrying the annotation specified.
Some examples of the use of this tool:
- Extract all tRNA gene sequences from a genome.
- Extract sequences for regions where annotations contain particular text
- Extract sequences of differentially expressed regions by using RNA-Seq statistical comparisons as the annotation source.
To launch Extract Annotated Regions, go to:
Toolbox | Utility Tools (![]() ) | Extract Annotated Regions (
) | Extract Annotated Regions (![]() )
)
This opens the wizard. In the first step (figure 35.1) you can select one or more annotated sequences, annotation tracks, variant tracks, or statistical comparison tracks.
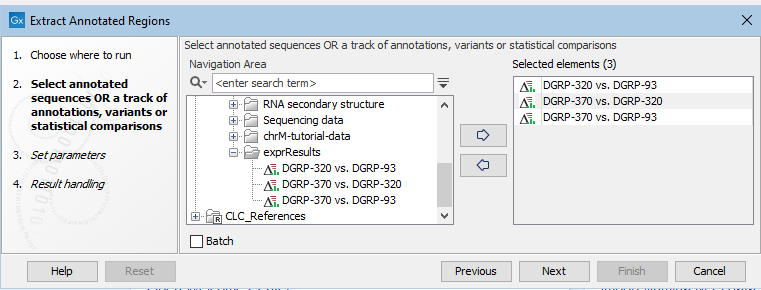
Figure 35.1: Selecting input. Here, statistical comparisontracks have been selected.
If you selected tracks as input, you must enter a reference sequence track in the next wizard step. In that step, you can also specify particular annotations where regions should be extracted, and flanking region lengths to be included, if desired (figure 35.2).
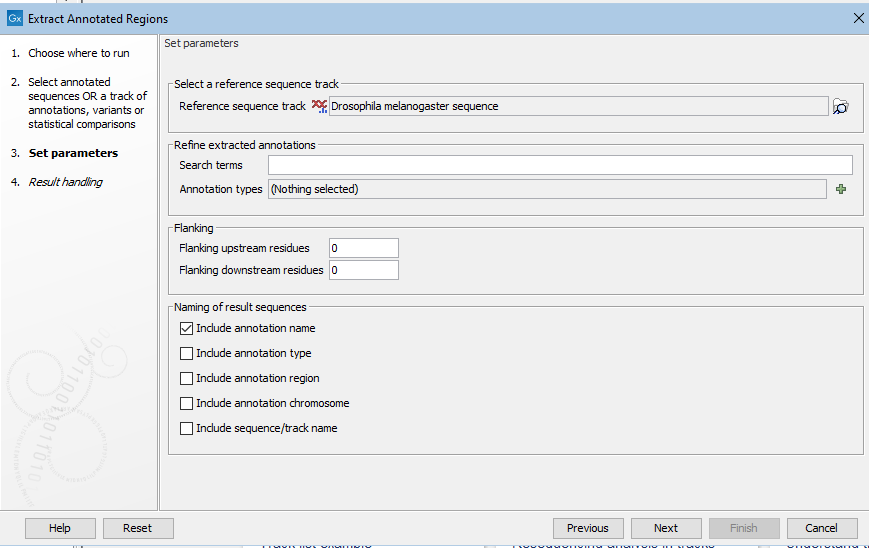
Figure 35.2: Specifying a reference sequence track after track-based data was selected as input.
- Search terms All annotations and attached information for each annotation will be searched for the entered term. It can be used to make general searches for search terms such as "Gene" or "Exon", or it can be used to make more specific searches. For example, if you have a gene annotation called "MLH1" and another called "MLH3", you can extract both annotations by entering "MLH" in the search term field. If you wish to enter more specific search terms, separate them with commas: "MLH1, Human" will find annotations where both "MLH1" and "Human" are included.
- Annotation types If only certain types of annotations should be extracted, this can be specified here.
The sequence of interest can be extracted with flanking sequences:
- Flanking upstream residues The output will include this number of extra residues at the 5' end of the annotation.
- Flanking downstream residues The output will include this number of extra residues at the 3' end of the annotation.
The sequences that are created can be named after the annotation name, type, etc:
- Include annotation name This will use the name of the annotation in the name of the extracted sequence.
- Include annotation type This corresponds to the type chosen above and will put this information in the name of the resulting sequences. This is useful information if you have chosen to extract "All" types of annotations.
- Include annotation region The region covered by the annotation on the original sequence (i.e. not including flanking regions) will be included in the name.
- Include sequence/track name If you have selected more than one sequence as input, this option enables you to discern the origin of the resulting sequences in the list by putting the name of the original sequence into the name of the resulting sequences.
Click Finish to start the tool.
