Standard import
CLC Genomics Workbench has support for a wide range of bioinformatic data formats. See List of supported data formats for a list.
For import of NGS data, please see Import high-throughput sequencing data For import of tracks, please see Import tracks.
The Standard Import tool
To import data, click on the Import (![]() ) icon in the Toolbar and choose Standard Import.
) icon in the Toolbar and choose Standard Import.
Alternatively, go to File | Import (![]() ) | Standard Import (
) | Standard Import (![]() ).
).
Select files to import by clicking on the Add files button. Alternatively, specify folders containing files to import by clicking on the Add folders button (figure 7.1).
The default option is Automatic import. The file format is automatically detected based on a combination of the file extension (e.g. .fa for fasta) and detection of file contents specific to particular formats. Based on this, the relevant importer is run. The particular importer used is recorded in the element history. If the format is not supported, the file is imported as an external file.
Note: If you drag and drop files into the Navigation Area, standard import is run with automatic detection of the file format.
You can specify the format explicitly using the option Force import as type.
The Force import as external file option can be useful if you are trying to import a standard format file, such as a text file, but it is being detected as bioinformatics format file, such as sequence data.
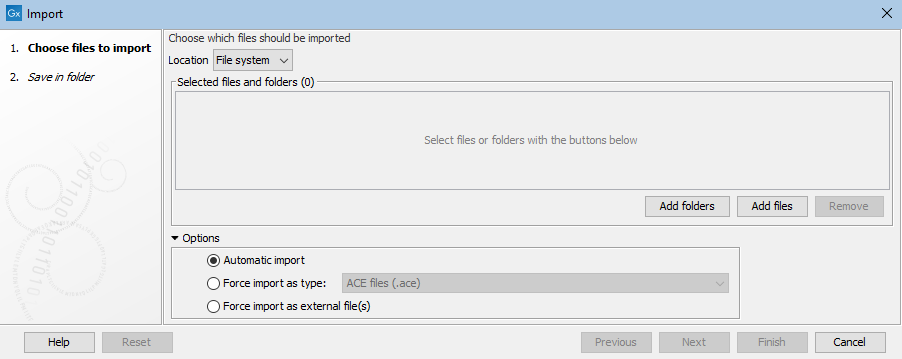
Figure 7.1: The Standard Import tool.
Importing by copy/pasting text
Data can be copied and pasted directly into the Navigation Area.
Copy text |
Select a folder in the Navigation Area |
Paste (![]() )
)
Standard import with automatic format detection is run using the pasted content as input.
This is a fast way to import data, but importing files as described above is less error prone, and thus generally recommended instead.
Subsections
