Sequencing primers
This mode is used to design primers for DNA sequencing.
In this mode the user can define a number of Forward primer regions and Reverse primer regions where a sequencing primer can start. These are defined by making a selection on the sequence and right-clicking the selection. If areas are known where primers must not bind (e.g. repeat rich areas), one or more No primers here regions can be defined.
No requirements are instated on the relative position of the regions defined.
After exploring the available primers (see Graphical display of primer information) and setting the desired parameter values in the Primer Parameters preference group, the Calculate button will activate the primer design algorithm.
After pressing the Calculate button a dialog will appear (see figure 19.11).
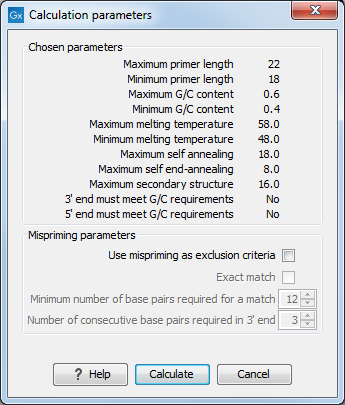
Figure 19.11: Calculation dialog for sequencing primers.
Since design of sequencing primers does not require the consideration of interactions between primer pairs, this dialog is identical to the dialog shown in Standard PCR mode when only a single primer region is chosen (see Standard PCR for a description).
Sequencing primers output table In this mode primers are predicted independently for each region, but the optimal solutions are all presented in one table. The solutions are numbered consecutively according to their position on the sequence such that the forward primer region closest to the 5' end of the molecule is designated F1, the next one F2 etc.
For each solution, the single primer information described under Standard PCR is available in the table.
